Stress-activated signalling pathways: starvation test 1
| Source | ID | Pathway |
|---|---|---|
| PathWhiz | PW003483 |
Pathway Metabolites
| YMDB ID | Name CAS Number | IUPAC Name | Formula Weight | Structure |
|---|---|---|---|---|
| YMDB00907 | phosphate 14265-44-2 | phosphoric acid | O4P 94.9714 | 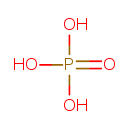 |
Pathway Proteins
| Uniprot ID | Gene Name Locus | Name | Type | Metabolites |
|---|---|---|---|---|
| P06784 | STE7 | Serine/threonine-protein kinase STE7 | Enzyme | Not Available |
| P08640 | FLO11 | Flocculation protein FLO11 | Not Available | |
| P11433 | CDC24 | Cell division control protein 24 | Not Available | |
| P13574 | STE12 | Protein STE12 | Not Available | |
| P14681 | KSS1 | Mitogen-activated protein kinase KSS1 | Enzyme | Not Available |
| P18412 | TEC1 | Ty transcription activator TEC1 | Not Available | |
| P19073 | CDC42 | Cell division control protein 42 | Not Available | |
| P23561 | STE11 | Serine/threonine-protein kinase STE11 | Enzyme | Not Available |
| P25344 | STE50 | Protein STE50 | Not Available | |
| P29311 | BMH1 | Protein BMH1 | Not Available | |
| P29461 | PTP2 | Tyrosine-protein phosphatase 2 | Enzyme | Not Available |
| P32329 | YPS1 | Aspartic proteinase 3 | Enzyme | Not Available |
| P32334 | MSB2 | Signaling mucin MSB2 | Not Available | |
| P34730 | BMH2 | Protein BMH2 | Not Available | |
| P38590 | MSG5 | Tyrosine-protein phosphatase MSG5 | Enzyme | Not Available |
| P40048 | PTP3 | Tyrosine-protein phosphatase 3 | Enzyme | Not Available |
| P40073 | SHO1 | High osmolarity signaling protein SHO1 | Not Available | |
| P53379 | MKC7 | Aspartic proteinase MKC7 | Enzyme | Not Available |
| Q03063 | DIG1 | Down-regulator of invasive growth 1 | Not Available | |
| Q03373 | DIG2 | Down-regulator of invasive growth 2 | Not Available | |
| Q03497 | STE20 | Serine/threonine-protein kinase STE20 | Enzyme | Not Available |
| Q03825 | MSS11 | Transcription activator MSS11 | Not Available | |
| Q06810 | OPY2 | Protein OPY2 | Not Available |