| Identification |
|---|
| Name | Tyrosine-protein phosphatase 2 |
|---|
| Synonyms | - Protein-tyrosine phosphatase 2
- PTPase 2
|
|---|
| Gene Name | PTP2 |
|---|
| Enzyme Class | Not Available |
|---|
| Biological Properties |
|---|
| General Function | regulation of sporulation |
|---|
| Specific Function | Major phosphatase responsible with PTP3 for tyrosine dephosphorylation of MAP kinase HOG1 to inactivate its activity. May also be involved in the regulation of MAP kinase FUS3. May be implicated in the ubiquitin-mediated protein degradation. |
|---|
| Cellular Location | Not Available |
|---|
| SMPDB Pathways | | Stress-activated signalling pathways: cell wall stress test 1 | PW007861 | 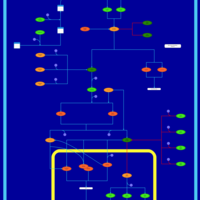 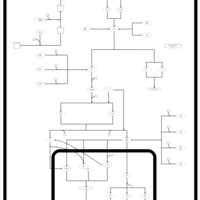 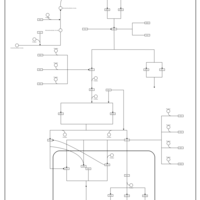 | | Stress-activated signalling pathways: high osmolarity test 1 | PW002773 | 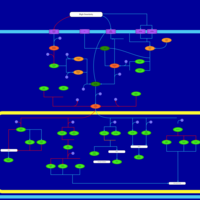 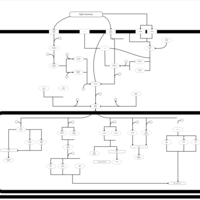 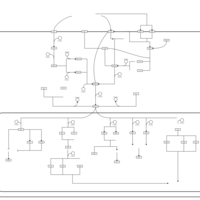 | | Stress-activated signalling pathways: low osmolarity | PW002515 |   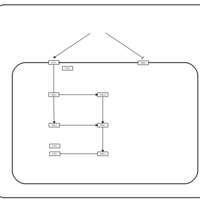 | | Stress-activated signalling pathways: pheromone stress test 1 | PW012843 |  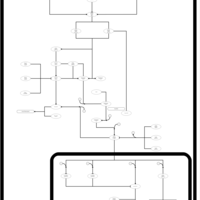 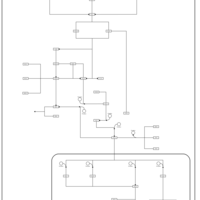 | | Stress-activated signalling pathways: starvation test 1 | PW003483 | 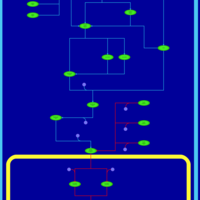 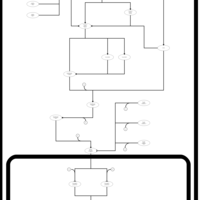 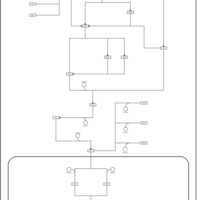 |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Metabolites | |
|---|
| GO Classification | | Function |
|---|
| nucleus | | cytoplasm | | regulation of protein localization | | protein tyrosine phosphatase activity | | protein dephosphorylation | | inactivation of MAPK activity involved in cell wall organization or biogenesis | | inactivation of MAPK activity involved in osmosensory signaling pathway | | regulation of sporulation |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | Not Available |
|---|
| Locus | |
|---|
| Gene Sequence | >2253 bp
ATGGATCGCATAGCACAGCAATATCGTAATGGCAAAAGAGACAATAACGGCAATAGAATG
GCTTCTTCCGCTATATCGGAAAAGGGCCACATACAAGTCAATCAAACTAGAACACCTGGT
CAAATGCCCGTCTATAGAGGTGAAACTATAAATCTGTCTAACCTTCCCCAAAATCAAATT
AAACCGTGCAAAGATTTGGACGACGTTAACATACGGCGGAACAACTCTAATAGGCATTCT
AAAATACTTTTACTAGATCTGTGCGCTGGCCCCAATACCAACTCATTTTTAGGCAATACC
AATGCTAAGGATATCACAGTTTTATCGTTGCCGCTACCCAGCACTTTGGTGAAAAGGTCG
AACTACCCGTTCGAGAACTTACTAAAGAATTACCTTGGATCTGATGAAAAGTATATTGAG
TTCACAAAGATCATCAAAGATTATGATATTTTCATTTTCAGTGATTCGTTTAGCAGAATT
TCGAGTTGTTTAAAGACAACTTTTTGCCTCATTGAGAAGTTTAAAAAGTTCATCTGCCAT
TTTTTTCCATCTCCTTATTTGAAATTCTTTCTTCTCGAAGGCTCTCTGAATGATAGCAAG
GCCCCCTCATTAGGAAAAAATAAGAAAAATTGCATCTTGCCCAAATTGGATTTGAACTTG
AATGTAAACTTAACTTCAAGGTCAACTTTAAATTTAAGAATAAACATACCTCCACCCAAT
GATTCAAATAAAATATTTTTACAGTCTCTGAAAAAGGATCTAATTCATTATTCTCCTAAT
TCTTTGCAAAAGTTTTTCCAATTCAATATGCCTGCTGACTTAGCACCTAACGACACGATT
TTACCGAATTGGCTAAAATTCTGCTCCGTAAAAGAAAATGAAAAGGTAATATTAAAGAAA
CTCTTTAACAATTTTGAAACTTTAGAAAATTTTGAAATGCAAAGATTAGAGAAATGCCTG
AAATTCAAGAAAAAGCCTTTACATCAAAAGCAGCTATCACAAAAGCAGAGGGGTCCGCAA
TCCACGGATGATTCAAAATTATATTCTTTAACTAGTTTGCAACGACAGTATAAAAGTTCT
TTGAAAAGCAACATACAGAAAAATCAAAAGCTAAAATTAATTATACCAAAAAACAACACA
TCTTCTTCGCCATCACCATTATCTTCCGATGATACTATAATGTCACCAATAAATGATTAC
GAACTTACTGAAGGAATTCAGTCTTTTACTAAGAATAGATATTCTAATATCTTACCTTAC
GAACATTCAAGAGTAAAGTTACCTCACTCCCCGAAACCACCTGCAGTTTCTGAAGCATCC
ACAACCGAAACTAAAACAGATAAGTCATATCCGATGTGTCCCGTAGATGCAAAAAACCAC
TCCTGCAAACCGAACGACTATATCAATGCGAACTATTTGAAGCTCACGCAAATTAATCCT
GATTTCAAGTATATTGCTACCCAAGCTCCGCTTCCTTCTACGATGGATGATTTTTGGAAG
GTTATTACTTTAAATAAAGTTAAAGTAATAATATCATTGAATTCTGACGATGAATTGAAT
TTAAGAAAATGGGATATTTACTGGAATAATCTGTCATATTCCAACCACACTATCAAACTT
CAGAACACCTGGGAGAATATTTGCAATATTAATGGCTGTGTTCTCAGAGTCTTTCAAGTC
AAGAAAACAGCTCCACAAAATGATAATATCAGTCAAGATTGTGACCTTCCGCATAATGGT
GACCTTACTTCCATTACCATGGCTGTATCCGAGCCGTTTATTGTTTACCAATTACAATAC
AAGAATTGGTTAGATTCATGCGGCGTAGATATGAATGACATCATTAAACTACACAAAGTC
AAAAATTCGTTATTGTTTAACCCGCAAAGTTTTATTACAAGCCTCGAAAAGGATGTTTGC
AAGCCTGATTTGATAGATGATAATAATAGTGAGTTACATCTCGATACAGCAAATTCATCG
CCACTATTAGTCCATTGTTCTGCAGGGTGTGGAAGAACAGGTGTTTTCGTTACCTTGGAT
TTCCTACTAAGTATTCTTTCACCTACAACAAATCACTCAAACAAGATTGATGTTTGGAAT
ATGACTCAGGACCTTATCTTTATCATAGTGAATGAATTAAGAAAGCAAAGGATTTCAATG
GTACAGAATCTAACTCAATATATCGCTTGTTATGAGGCATTATTAAATTATTTTGCCCTG
CAAAAGCAGATAAAGAACGCGTTACCTTGTTAA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | Not Available |
|---|
| Protein Residues | 750 |
|---|
| Protein Molecular Weight | 85867.605 |
|---|
| Protein Theoretical pI | Not Available |
|---|
| Signalling Regions | Not Available |
|---|
| Transmembrane Regions | Not Available |
|---|
| Protein Sequence | >Tyrosine-protein phosphatase 2
MDRIAQQYRNGKRDNNGNRMASSAISEKGHIQVNQTRTPGQMPVYRGETINLSNLPQNQI
KPCKDLDDVNIRRNNSNRHSKILLLDLCAGPNTNSFLGNTNAKDITVLSLPLPSTLVKRS
NYPFENLLKNYLGSDEKYIEFTKIIKDYDIFIFSDSFSRISSCLKTTFCLIEKFKKFICH
FFPSPYLKFFLLEGSLNDSKAPSLGKNKKNCILPKLDLNLNVNLTSRSTLNLRINIPPPN
DSNKIFLQSLKKDLIHYSPNSLQKFFQFNMPADLAPNDTILPNWLKFCSVKENEKVILKK
LFNNFETLENFEMQRLEKCLKFKKKPLHQKQLSQKQRGPQSTDDSKLYSLTSLQRQYKSS
LKSNIQKNQKLKLIIPKNNTSSSPSPLSSDDTIMSPINDYELTEGIQSFTKNRYSNILPY
EHSRVKLPHSPKPPAVSEASTTETKTDKSYPMCPVDAKNHSCKPNDYINANYLKLTQINP
DFKYIATQAPLPSTMDDFWKVITLNKVKVIISLNSDDELNLRKWDIYWNNLSYSNHTIKL
QNTWENICNINGCVLRVFQVKKTAPQNDNISQDCDLPHNGDLTSITMAVSEPFIVYQLQY
KNWLDSCGVDMNDIIKLHKVKNSLLFNPQSFITSLEKDVCKPDLIDDNNSELHLDTANSS
PLLVHCSAGCGRTGVFVTLDFLLSILSPTTNHSNKIDVWNMTQDLIFIIVNELRKQRISM
VQNLTQYIACYEALLNYFALQKQIKNALPC |
|---|
| References |
|---|
| External Links | | Resource | Link |
|---|
| Saccharomyces Genome Database | PTP2 | | Uniprot ID | P29461 | | Uniprot Name | PTP2 |
|
|---|
| General Reference | Not Available |
|---|