| Identification |
|---|
| Name | Aspartic proteinase 3 |
|---|
| Synonyms | - Proprotein convertase
- Yapsin-1
|
|---|
| Gene Name | YPS1 |
|---|
| Enzyme Class | Not Available |
|---|
| Biological Properties |
|---|
| General Function | signal transduction involved in filamentous growth |
|---|
| Specific Function | Cleaves proteins C-terminally to mono- and paired-basic residues. Involved in the shedding of a subset of GPI-anchored plasma membrane proteins from the cell surface, including itself, GAS1 and MSB2. May also play a role in the maturation of GPI-mannoproteins associated with the cell wall. Can process the alpha-mating factor precursor. Required for cell wall integrity. |
|---|
| Cellular Location | Not Available |
|---|
| SMPDB Pathways | | Stress-activated signalling pathways: starvation test 1 | PW003483 | 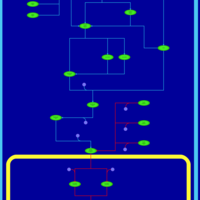   |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Metabolites | |
|---|
| GO Classification | | Function |
|---|
| fungal-type cell wall organization | | anchored component of plasma membrane | | peptide mating pheromone maturation involved in conjugation with cellular fusion | | protein processing | | signal transduction involved in filamentous growth | | aspartic-type endopeptidase activity | | protein catabolic process |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | Not Available |
|---|
| Locus | |
|---|
| Gene Sequence | >1710 bp
ATGAAACTGAAAACTGTAAGATCTGCGGTCCTTTCGTCACTCTTTGCATCGCAGGTTCTC
GGTAAGATAATACCAGCAGCAAACAAGCGCGACGACGACTCGAATTCCAAGTTCGTCAAG
TTGCCCTTTCATAAGCTTTACGGGGACTCGCTAGAAAATGTGGGAAGCGACAAAAAACCG
GAAGTACGCCTATTGAAGAGGGCTGACGGTTATGAAGAAATTATAATTACCAACCAGCAA
AGTTTCTATTCGGTGGACTTGGAAGTGGGCACGCCACCACAGAACGTAACGGTCCTGGTG
GACACAGGCTCCTCTGATCTATGGATTATGGGCTCGGATAATCCATACTGTTCTTCGAAC
AGTATGGGTAGTAGCCGGAGACGTGTTATTGACAAACGTGATGATTCGTCAAGCGGCGGA
TCTTTGATTAATGATATAAACCCATTTGGCTGGTTGACGGGAACGGGCAGTGCCATTGGC
CCCACTGCTACGGGCTTAGGAGGCGGTTCAGGTACGGCAACTCAATCCGTGCCTGCTTCG
GAAGCCACCATGGACTGTCAACAATACGGGACATTTTCCACTTCGGGCTCTTCTACATTT
AGATCAAACAACACCTATTTCAGTATTAGCTACGGTGATGGGACTTTTGCCTCCGGTACT
TTTGGTACGGATGTTTTGGATTTAAGCGACTTGAACGTTACCGGGTTGTCTTTTGCCGTT
GCCAATGAAACGAATTCTACTATGGGTGTGTTAGGTATTGGTTTGCCCGAATTAGAAGTC
ACTTATTCTGGCTCTACTGCGTCTCATAGTGGAAAAGCTTATAAATACGACAACTTCCCC
ATTGTATTGAAAAATTCTGGTGCTATCAAAAGCAACACATATTCTTTGTATTTGAACGAC
TCGGACGCTATGCATGGCACCATTTTGTTCGGAGCCGTGGACCACAGTAAATATACCGGC
ACCTTATACACAATCCCCATCGTAAACACTCTGAGTGCTAGTGGATTTAGCTCTCCCATT
CAATTTGATGTCACTATTAATGGTATCGGTATTAGTGATTCTGGGAGTAGTAACAAGACC
TTGACTACCACTAAAATACCTGCTTTGTTGGATTCCGGTACTACTTTGACTTATTTACCT
CAAACAGTGGTAAGTATGATCGCTACTGAACTAGGTGCGCAATACTCTTCCAGGATAGGG
TATTACGTATTGGACTGTCCATCTGATGATAGTATGGAAATAGTGTTCGATTTTGGTGGT
TTTCACATCAATGCACCACTTTCGAGTTTTATCTTGAGTACTGGCACTACATGTCTTTTA
GGTATTATCCCAACGAGTGATGACACAGGTACCATTTTGGGTGATTCATTTTTGACTAAC
GCGTACGTGGTTTATGATTTGGAGAATCTTGAAATATCCATGGCACAAGCTCGCTATAAT
ACCACAAGCGAAAATATCGAAATTATTACATCCTCTGTTCCAAGCGCCGTAAAGGCACCA
GGCTATACAAACACTTGGTCCACAAGTGCATCTATTGTTACCGGTGGTAACATATTTACT
GTAAATTCCTCACAAACTGCTTCCTTTAGCGGTAACCTGACGACCAGTACTGCATCCGCC
ACTTCTACATCAAGTAAAAGAAATGTTGGTGATCATATAGTTCCATCTTTACCCCTCACA
TTAATTTCTCTTCTTTTTGCATTCATCTGA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | Not Available |
|---|
| Protein Residues | 569 |
|---|
| Protein Molecular Weight | 60009.13 |
|---|
| Protein Theoretical pI | Not Available |
|---|
| PDB File | show |
|---|
| Signalling Regions | |
|---|
| Transmembrane Regions | Not Available |
|---|
| Protein Sequence | >Aspartic proteinase 3
MKLKTVRSAVLSSLFASQVLGKIIPAANKRDDDSNSKFVKLPFHKLYGDSLENVGSDKKP
EVRLLKRADGYEEIIITNQQSFYSVDLEVGTPPQNVTVLVDTGSSDLWIMGSDNPYCSSN
SMGSSRRRVIDKRDDSSSGGSLINDINPFGWLTGTGSAIGPTATGLGGGSGTATQSVPAS
EATMDCQQYGTFSTSGSSTFRSNNTYFSISYGDGTFASGTFGTDVLDLSDLNVTGLSFAV
ANETNSTMGVLGIGLPELEVTYSGSTASHSGKAYKYDNFPIVLKNSGAIKSNTYSLYLND
SDAMHGTILFGAVDHSKYTGTLYTIPIVNTLSASGFSSPIQFDVTINGIGISDSGSSNKT
LTTTKIPALLDSGTTLTYLPQTVVSMIATELGAQYSSRIGYYVLDCPSDDSMEIVFDFGG
FHINAPLSSFILSTGTTCLLGIIPTSDDTGTILGDSFLTNAYVVYDLENLEISMAQARYN
TTSENIEIITSSVPSAVKAPGYTNTWSTSASIVTGGNIFTVNSSQTASFSGNLTTSTASA
TSTSSKRNVGDHIVPSLPLTLISLLFAFI |
|---|
| References |
|---|
| External Links | | Resource | Link |
|---|
| Saccharomyces Genome Database | YPS1 | | Uniprot ID | P32329 | | Uniprot Name | YPS1 | | PDB ID | 1YPS |
|
|---|
| General Reference | Not Available |
|---|