Cyanoamino acid metabolism
| Source | ID | Pathway |
|---|---|---|
| KEGG | 00460 | 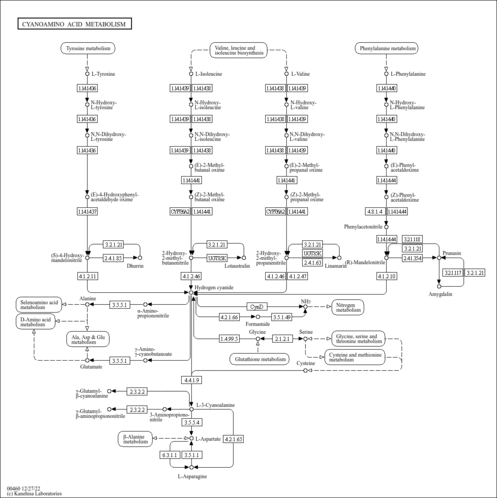 |
Pathway Metabolites
| YMDB ID | Name CAS Number | IUPAC Name | Formula Weight | Structure |
|---|---|---|---|---|
| YMDB00016 | Glycine 56-40-6 | 2-aminoacetic acid | C2H5NO2 75.0666 |  |
| YMDB00074 | Cyanide 57-12-5 | methylidyneazanidyl | CN 26.0174 |  |
| YMDB00091 | Ammonia 7664-41-7 | ammonia | H3N 17.0305 |  |
| YMDB00112 | L-Serine 56-45-1 | (2S)-2-amino-3-hydroxypropanoic acid | C3H7NO3 105.0926 | 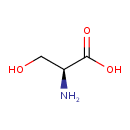 |
| YMDB00226 | L-Asparagine 70-47-3 | (2S)-2-amino-3-carbamoylpropanoic acid | C4H8N2O3 132.1179 | 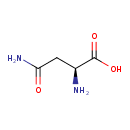 |
| YMDB00364 | L-Tyrosine 60-18-4 | (2S)-2-amino-3-(4-hydroxyphenyl)propanoic acid | C9H11NO3 181.1885 | 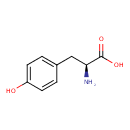 |
| YMDB00784 | gamma-amino-gamma-cyanobutanoic acid 14046-56-1 | 4-amino-4-cyanobutanoic acid | C5H8N2O2 128.1292 | 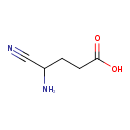 |
| YMDB00896 | L-Aspartic acid 56-84-8 | (2S)-2-aminobutanedioic acid | C4H7NO4 133.1027 | 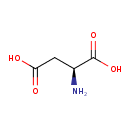 |
| YMDB00912 | Carbon dioxide 124-38-9 | methanedione | CO2 44.0095 | 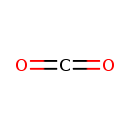 |
Pathway Proteins
| Uniprot ID | Gene Name Locus | Name | Type | Metabolites |
|---|---|---|---|---|
| P37291 | SHM2 YLR058C | Serine hydroxymethyltransferase, cytosolic | Enzyme | |
| P37292 | SHM1 YBR263W | Serine hydroxymethyltransferase, mitochondrial | Enzyme | |
| P11163 | ASP3-1 YLR155C | L-asparaginase 2 | Enzyme | |
| P38986 | ASP1 YDR321W | L-asparaginase 1 | Enzyme | |
| P22580 | AMD2 YDR242W | Probable amidase | Enzyme | |
| Q05902 | ECM38 YLR299W | Gamma-glutamyltransferase | Enzyme |