| Identification |
|---|
| Name | Peroxisomal hydratase-dehydrogenase-epimerase |
|---|
| Synonyms | - HDE
- Multifunctional beta-oxidation protein
- MFP
- 2-enoyl-CoA hydratase
- (3R)-3-hydroxyacyl-CoA dehydrogenase
|
|---|
| Gene Name | FOX2 |
|---|
| Enzyme Class | |
|---|
| Biological Properties |
|---|
| General Function | Involved in oxidoreductase activity |
|---|
| Specific Function | Second trifunctional enzyme acting on the beta-oxidation pathway for fatty acids, possessing hydratase-dehydrogenase- epimerase activities. Converts trans-2-enoyl-CoA via D-3- hydroxyacyl-CoA to 3-ketoacyl-CoA |
|---|
| Cellular Location | Peroxisome |
|---|
| SMPDB Pathways | | Biosynthesis of unsaturated fatty acids | PW002403 |  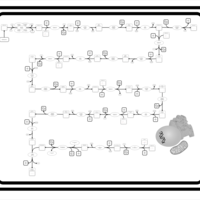 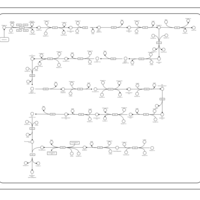 |
|
|---|
| KEGG Pathways | | Biosynthesis of unsaturated fatty acids | ec01040 |  |
|
|---|
| SMPDB Reactions |
|
|---|
| KEGG Reactions |
|
|---|
| Metabolites | | YMDB ID | Name | View |
|---|
| YMDB00110 | NAD | Show | | YMDB00143 | NADH | Show | | YMDB00412 | NAD(+) | Show | | YMDB00463 | trans-dec-2-enoyl-CoA | Show | | YMDB00464 | (S)-3-hydroxydecanoyl-CoA | Show | | YMDB00465 | (S)-3-hydroxylauroyl-CoA | Show | | YMDB00466 | trans-dodec-2-enoyl-CoA | Show | | YMDB00467 | trans-hexacos-2-enoyl-CoA | Show | | YMDB00468 | (S)-3-hydroxypalmitoyl-CoA | Show | | YMDB00469 | hexadec-2-enoyl-CoA | Show | | YMDB00470 | trans-octadec-2-enoyl-CoA | Show | | YMDB00471 | 3-hydroxyoctadecanoyl-CoA | Show | | YMDB00472 | trans-tetradec-2-enoyl-CoA | Show | | YMDB00473 | (S)-3-hydroxytetradecanoyl-CoA | Show | | YMDB00474 | 3-oxodecanoyl-CoA | Show | | YMDB00475 | 3-oxolauroyl-CoA | Show | | YMDB00476 | 3-oxopalmitoyl-CoA | Show | | YMDB00477 | 3-oxooctadecanoyl-CoA | Show | | YMDB00478 | 3-oxotetradecanoyl-CoA | Show | | YMDB00862 | hydron | Show | | YMDB00890 | water | Show | | YMDB00959 | (S)-3-hydroxyhexacosanoyl-CoA | Show | | YMDB00960 | 3-oxohexacosanoyl-CoA | Show | | YMDB16167 | (S)-Hydroxyoctanoyl-CoA | Show | | YMDB16168 | 3-Oxooctanoyl-CoA | Show | | YMDB16169 | (S)-Hydroxyhexanoyl-CoA | Show | | YMDB16170 | 3-Oxohexanoyl-CoA | Show | | YMDB16292 | (3R)-3-hydroxy-stearoyl-CoA | Show | | YMDB16293 | (3R)-3-hydroxy-arachidoyl-CoA | Show | | YMDB16294 | (2E)-eicosenoyl-CoA | Show | | YMDB16295 | (3R)-3-hydroxy-docosanoyl-CoA | Show | | YMDB16296 | trans-docos-2-enoyl-CoA | Show | | YMDB16297 | (3R)-hydroxytetracosanoyl-CoA | Show | | YMDB16298 | (2E)-lignocerenoyl-CoA | Show | | YMDB16299 | (3R)-hydroxyhexacosanoyl-CoA | Show | | YMDB16300 | (2E)-cerotenoyl-CoA | Show |
|
|---|
| GO Classification | | Component |
|---|
| Not Available | | Function |
|---|
| catalytic activity | | binding | | oxidoreductase activity | | Process |
|---|
| metabolic process | | oxidation reduction |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | chromosome 11 |
|---|
| Locus | YKR009C |
|---|
| Gene Sequence | >2703 bp
ATGCCTGGAAATTTATCCTTCAAAGATAGAGTTGTTGTAATCACGGGCGCTGGAGGGGGC
TTAGGTAAGGTGTATGCACTAGCTTACGCAAGCAGAGGTGCAAAAGTGGTCGTCAATGAT
CTAGGTGGCACTTTGGGTGGTTCAGGACATAACTCCAAAGCTGCAGACTTAGTGGTGGAT
GAGATAAAAAAAGCCGGAGGTATAGCTGTGGCAAATTACGACTCTGTTAATGAAAATGGA
GAGAAAATAATTGAAACGGCTATAAAAGAATTCGGCAGGGTTGATGTACTAATTAACAAC
GCTGGAATATTAAGGGATGTTTCATTTGCAAAGATGACAGAACGTGAGTTTGCATCTGTG
GTAGATGTTCATTTGACAGGTGGCTATAAGCTATCGCGTGCTGCTTGGCCTTATATGCGC
TCTCAGAAATTTGGTAGAATCATTAACACCGCTTCCCCTGCCGGTCTATTTGGAAATTTT
GGTCAAGCTAATTATTCAGCAGCTAAAATGGGCTTAGTTGGTTTGGCGGAAACCCTCGCG
AAGGAGGGTGCCAAATACAACATTAATGTTAATTCAATTGCGCCATTGGCTAGATCACGT
ATGACAGAAAACGTGTTACCACCACATATCTTGAAACAGTTAGGACCGGAAAAAATTGTT
CCCTTAGTACTCTATTTGACACACGAAAGTACGAAAGTGTCAAACTCCATTTTTGAACTC
GCTGCTGGATTCTTTGGACAGCTCAGATGGGAGAGGTCTTCTGGACAAATTTTCAATCCA
GACCCCAAGACATATACTCCTGAAGCAATTTTAAATAAGTGGAAGGAAATCACAGACTAT
AGGGACAAGCCATTTAACAAAACTCAGCATCCATATCAACTCTCGGATTATAATGATTTA
ATCACCAAAGCAAAAAAATTACCTCCCAATGAACAAGGCTCAGTGAAAATCAAGTCGCTT
TGCAACAAAGTCGTAGTAGTTACGGGTGCAGGAGGTGGTCTTGGGAAGTCTCATGCAATC
TGGTTTGCACGGTACGGTGCGAAGGTAGTTGTAAATGACATCAAGGATCCTTTTTCAGTT
GTTGAAGAAATAAATAAACTATATGGTGAAGGCACAGCCATTCCAGATTCCCATGATGTG
GTCACCGAAGCTCCTCTCATTATCCAAACTGCAATAAGTAAGTTTCAGAGAGTAGACATC
TTGGTCAATAACGCTGGTATTTTGCGTGACAAATCTTTTTTAAAAATGAAAGATGAGGAA
TGGTTTGCTGTCCTGAAAGTCCACCTTTTTTCCACATTTTCATTGTCAAAAGCAGTATGG
CCAATATTTACCAAACAAAAGTCTGGATTTATTATCAATACTACTTCTACCTCAGGAATT
TATGGTAATTTTGGACAGGCCAATTATGCCGCTGCAAAAGCCGCCATTTTAGGATTCAGT
AAAACTATTGCACTGGAAGGTGCCAAGAGAGGAATTATTGTTAATGTTATCGCTCCTCAT
GCAGAAACGGCTATGACAAAGACTATATTCTCGGAGAAGGAATTATCAAACCACTTTGAT
GCATCTCAAGTCTCCCCACTTGTTGTTTTGTTGGCATCTGAAGAACTACAAAAGTATTCT
GGAAGAAGGGTTATTGGCCAATTATTCGAAGTTGGCGGTGGTTGGTGTGGGCAAACCAGA
TGGCAAAGAAGTTCCGGTTATGTTTCTATTAAAGAGACTATTGAACCGGAAGAAATTAAA
GAAAATTGGAACCACATCACTGATTTCAGTCGCAACACTATCAACCCGAGCTCCACAGAG
GAGTCTTCTATGGCAACCTTGCAAGCCGTGCAAAAAGCGCACTCTTCAAAGGAGTTGGAT
GATGGATTATTCAAGTACACTACCAAGGATTGTATCTTGTACAATTTAGGACTTGGATGC
ACAAGCAAAGAGCTTAAGTACACCTACGAGAATGATCCAGACTTCCAAGTTTTGCCCACG
TTCGCCGTCATTCCATTTATGCAAGCTACTGCCACACTAGCTATGGACAATTTAGTCGAT
AACTTCAATTATGCAATGTTACTGCATGGAGAACAATATTTTAAGCTCTGCACGCCGACA
ATGCCAAGTAATGGAACTCTAAAGACACTTGCTAAACCTTTACAAGTACTTGACAAGAAT
GGTAAAGCCGCTTTAGTTGTTGGTGGCTTCGAAACTTATGACATTAAAACTAAGAAACTC
ATAGCTTATAACGAAGGATCGTTCTTCATCAGGGGCGCACATGTACCTCCAGAAAAGGAA
GTGAGGGATGGGAAAAGAGCCAAGTTTGCTGTCCAAAATTTTGAAGTGCCACATGGAAAG
GTACCAGATTTTGAGGCGGAGATTTCTACGAATAAAGATCAAGCCGCATTGTACAGGTTA
TCTGGCGATTTCAATCCTTTACATATCGATCCCACGCTAGCCAAAGCAGTTAAATTTCCT
ACGCCAATTCTGCATGGGCTTTGTACATTAGGTATTAGTGCGAAAGCATTGTTTGAACAT
TATGGTCCATATGAGGAGTTGAAAGTGAGATTTACCAATGTTGTTTTCCCAGGTGATACT
CTAAAGGTTAAAGCTTGGAAGCAAGGCTCGGTTGTCGTTTTTCAAACAATTGATACGACC
AGAAACGTCATTGTATTGGATAACGCCGCTGTAAAACTATCGCAGGCAAAATCTAAACTA
TAA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | |
|---|
| Protein Residues | 900 |
|---|
| Protein Molecular Weight | 98702.39844 |
|---|
| Protein Theoretical pI | 9.46 |
|---|
| Signalling Regions | |
|---|
| Transmembrane Regions | |
|---|
| Protein Sequence | >Peroxisomal hydratase-dehydrogenase-epimerase
MPGNLSFKDRVVVITGAGGGLGKVYALAYASRGAKVVVNDLGGTLGGSGHNSKAADLVVD
EIKKAGGIAVANYDSVNENGEKIIETAIKEFGRVDVLINNAGILRDVSFAKMTEREFASV
VDVHLTGGYKLSRAAWPYMRSQKFGRIINTASPAGLFGNFGQANYSAAKMGLVGLAETLA
KEGAKYNINVNSIAPLARSRMTENVLPPHILKQLGPEKIVPLVLYLTHESTKVSNSIFEL
AAGFFGQLRWERSSGQIFNPDPKTYTPEAILNKWKEITDYRDKPFNKTQHPYQLSDYNDL
ITKAKKLPPNEQGSVKIKSLCNKVVVVTGAGGGLGKSHAIWFARYGAKVVVNDIKDPFSV
VEEINKLYGEGTAIPDSHDVVTEAPLIIQTAISKFQRVDILVNNAGILRDKSFLKMKDEE
WFAVLKVHLFSTFSLSKAVWPIFTKQKSGFIINTTSTSGIYGNFGQANYAAAKAAILGFS
KTIALEGAKRGIIVNVIAPHAETAMTKTIFSEKELSNHFDASQVSPLVVLLASEELQKYS
GRRVIGQLFEVGGGWCGQTRWQRSSGYVSIKETIEPEEIKENWNHITDFSRNTINPSSTE
ESSMATLQAVQKAHSSKELDDGLFKYTTKDCILYNLGLGCTSKELKYTYENDPDFQVLPT
FAVIPFMQATATLAMDNLVDNFNYAMLLHGEQYFKLCTPTMPSNGTLKTLAKPLQVLDKN
GKAALVVGGFETYDIKTKKLIAYNEGSFFIRGAHVPPEKEVRDGKRAKFAVQNFEVPHGK
VPDFEAEISTNKDQAALYRLSGDFNPLHIDPTLAKAVKFPTPILHGLCTLGISAKALFEH
YGPYEELKVRFTNVVFPGDTLKVKAWKQGSVVVFQTIDTTRNVIVLDNAAVKLSQAKSKL
|
|---|
| References |
|---|
| External Links | |
|---|
| General Reference | - Dujon, B., Alexandraki, D., Andre, B., Ansorge, W., Baladron, V., Ballesta, J. P., Banrevi, A., Bolle, P. A., Bolotin-Fukuhara, M., Bossier, P., et, a. l. .. (1994). "Complete DNA sequence of yeast chromosome XI." Nature 369:371-378.8196765
- Dusterhoft, A., Philippsen, P. (1992). "DNA sequencing and analysis of a 24.7 kb segment encompassing centromere CEN11 of Saccharomyces cerevisiae reveals nine previously unknown open reading frames." Yeast 8:749-759.1441752
- Hiltunen, J. K., Wenzel, B., Beyer, A., Erdmann, R., Fossa, A., Kunau, W. H. (1992). "Peroxisomal multifunctional beta-oxidation protein of Saccharomyces cerevisiae. Molecular analysis of the fox2 gene and gene product." J Biol Chem 267:6646-6653.1551874
|
|---|
