| Identification |
|---|
| Name | Catabolic L-serine/threonine dehydratase |
|---|
| Synonyms | - L-serine dehydratase
- L-serine deaminase
- L-threonine dehydratase
- L-threonine deaminase
|
|---|
| Gene Name | CHA1 |
|---|
| Enzyme Class | |
|---|
| Biological Properties |
|---|
| General Function | Involved in catalytic activity |
|---|
| Specific Function | L-serine = pyruvate + NH(3) |
|---|
| Cellular Location | Mitochondrion |
|---|
| SMPDB Pathways | |
|---|
| KEGG Pathways | | Glycine, serine and threonine metabolism | ec00260 |  | | Valine, leucine and isoleucine biosynthesis | ec00290 | 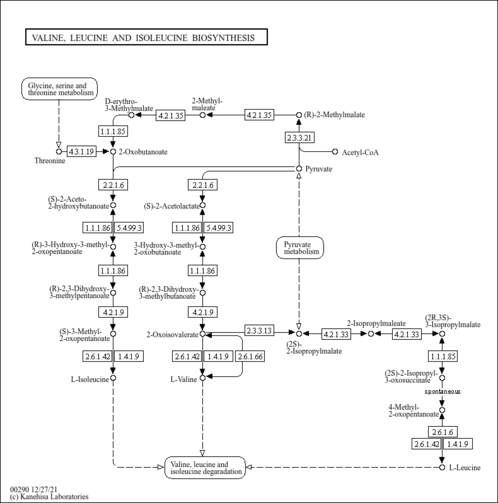 |
|
|---|
| SMPDB Reactions | |
|---|
| KEGG Reactions | |
|---|
| Metabolites | | YMDB ID | Name | View |
|---|
| YMDB00071 | 2-Ketobutyric acid | Show | | YMDB00091 | Ammonia | Show | | YMDB00112 | L-Serine | Show | | YMDB00175 | Pyruvic acid | Show | | YMDB00214 | L-Threonine | Show | | YMDB00423 | Ammonium | Show | | YMDB00862 | hydron | Show | | YMDB00890 | water | Show | | YMDB16258 | 2-aminoprop-2-enoate | Show |
|
|---|
| GO Classification | | Component |
|---|
| Not Available | | Function |
|---|
| catalytic activity | | binding | | cofactor binding | | pyridoxal phosphate binding | | Process |
|---|
| metabolic process | | cellular metabolic process | | cellular amino acid and derivative metabolic process | | cellular amino acid metabolic process |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | chromosome 3 |
|---|
| Locus | YCL064C |
|---|
| Gene Sequence | >1083 bp
ATGTCGATAGTCTACAATAAAACACCATTATTACGTCAATTCTTCCCCGGAAAGGCTTCT
GCACAATTTTTCTTGAAATATGAATGCCTTCAACCAAGTGCCTCCTTCAAAAGTAGAGGA
ATCGGTAATCTCATCATGAAAAGTGCCATTCGAATTCAAAAGGACGGTAAAAGATCTCCT
CAGGTTTTCGCTAGTTCTGGCGGTAATGCCGGTTTTGCTGCTGCAACAGCATGTCAAAGA
CTGTCTCTACCATGTACAGTCGTGGTTCCTACAGCGACAAAGAAGAGAATGGTAGATAAA
ATCAGGAACACCGGTGCCCAGGTTATCGTGAGTGGTGCCTACTGGAAAGAAGCAGATACT
TTTTTAAAAACAAATGTCATGAATAAAATAGACTCTCAGGTCATTGAGCCCATTTATGTT
CATCCCTTCGATAATCCGGATATTTGGGAAGGACATTCATCTATGATAGATGAAATAGTA
CAAGATTTGAAATCGCAACATATTTCCGTAAATAAGGTTAAAGGCATAGTATGCAGCGTT
GGTGGAGGTGGTTTATACAATGGTATTATTCAAGGTTTGGAAAGGTATGGTTTAGCTGAT
AGGATCCCTATTGTGGGGGTGGAAACGAATGGATGTCATGTTTTCAATACTTCTTTGAAA
ATAGGCCAACCAGTTCAATTCAAGAAGATAACAAGTATTGCTACTTCTCTAGGAACGGCC
GTGATCTCTAATCAAACTTTCGAATACGCTCGCAAATACAACACCAGATCCGTTGTAATA
GAGGACAAAGATGTGATTGAACCCTGTCTTAAATATACACATCAATTCAATATGGTGATT
GAACCGGCATGTGGCGCCGCATTGCATTTGGGTTACAACACTAAGATCCTAGAAAATGCA
CTGGGCTCAAAATTAGCTGCGGATGACATTGTGATAATTATTGCTTGTGCGAGCTCCTCT
AATACTATAAAGGACTTGGAAGAAGCGTTGGATAGCATGAGAAAAAAAGACACTCCTGTA
ATAGAAGTCGCTGACAATTTCATATTTCCAGAAAAAAATATTGTGAATTTAAAAAGTGCT
TGA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | |
|---|
| Protein Residues | 360 |
|---|
| Protein Molecular Weight | 39301.0 |
|---|
| Protein Theoretical pI | 9.01 |
|---|
| Signalling Regions | |
|---|
| Transmembrane Regions | |
|---|
| Protein Sequence | >Catabolic L-serine/threonine dehydratase
MSIVYNKTPLLRQFFPGKASAQFFLKYECLQPSGSFKSRGIGNLIMKSAIRIQKDGKRSP
QVFASSGGNAGFAAATACQRLSLPCTVVVPTATKKRMVDKIRNTGAQVIVSGAYWKEADT
FLKTNVMNKIDSQVIEPIYVHPFDNPDIWEGHSSMIDEIVQDLKSQHISVNKVKGIVCSV
GGGGLYNGIIQGLERYGLADRIPIVGVETNGCHVFNTSLKIGQPVQFKKITSIATSLGTA
VISNQTFEYARKYNTRSVVIEDKDVIETCLKYTHQFNMVIEPACGAALHLGYNTKILENA
LGSKLAADDIVIIIACGGSSNTIKDLEEALDSMRKKDTPVIEVADNFIFPEKNIVNLKSA
|
|---|
| References |
|---|
| External Links | |
|---|
| General Reference | - Bornaes, C., Petersen, J. G., Holmberg, S. (1992). "Serine and threonine catabolism in Saccharomyces cerevisiae: the CHA1 polypeptide is homologous with other serine and threonine dehydratases." Genetics 131:531-539.1628804
- Oliver, S. G., van der Aart, Q. J., Agostoni-Carbone, M. L., Aigle, M., Alberghina, L., Alexandraki, D., Antoine, G., Anwar, R., Ballesta, J. P., Benit, P., et, a. l. .. (1992). "The complete DNA sequence of yeast chromosome III." Nature 357:38-46.1574125
- Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., O'Shea, E. K. (2003). "Global analysis of protein localization in budding yeast." Nature 425:686-691.14562095
- Ghaemmaghami, S., Huh, W. K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., O'Shea, E. K., Weissman, J. S. (2003). "Global analysis of protein expression in yeast." Nature 425:737-741.14562106
|
|---|

