| Identification |
|---|
| Name | 5-aminolevulinate synthase, mitochondrial |
|---|
| Synonyms | - 5-aminolevulinic acid synthase
- Delta-ALA synthase
- Delta-aminolevulinate synthase
|
|---|
| Gene Name | HEM1 |
|---|
| Enzyme Class | |
|---|
| Biological Properties |
|---|
| General Function | Involved in 5-aminolevulinate synthase activity |
|---|
| Specific Function | Succinyl-CoA + glycine = 5-aminolevulinate + CoA + CO(2) |
|---|
| Cellular Location | Mitochondrion matrix |
|---|
| SMPDB Pathways | | Biosynthesis of unsaturated fatty acids (docosanoyl) | PW002408 | 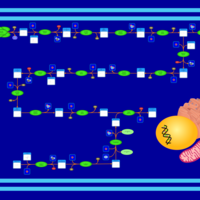 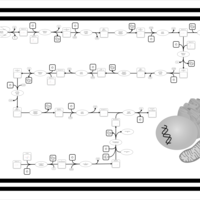 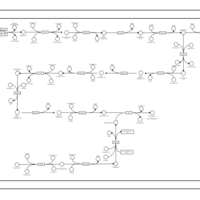 | | Biosynthesis of unsaturated fatty acids (tetracosanoyl-CoA) | PW002404 | 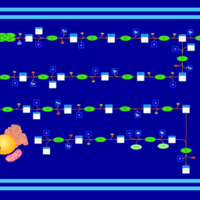 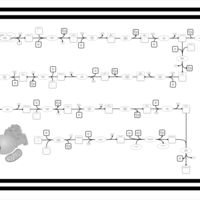 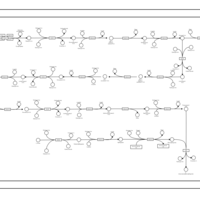 |
|
|---|
| KEGG Pathways | | Glycine, serine and threonine metabolism | ec00260 |  | | Porphyrin and chlorophyll metabolism | ec00860 |  |
|
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | |
|---|
| Metabolites | | YMDB ID | Name | View |
|---|
| YMDB00016 | Glycine | Show | | YMDB00045 | Coenzyme A | Show | | YMDB00083 | 5-Aminolevulinic acid | Show | | YMDB00514 | Succinyl-CoA | Show | | YMDB00862 | hydron | Show | | YMDB00912 | Carbon dioxide | Show |
|
|---|
| GO Classification | | Component |
|---|
| Not Available | | Function |
|---|
| catalytic activity | | transferase activity | | binding | | transferase activity, transferring nitrogenous groups | | cofactor binding | | pyridoxal phosphate binding | | transferase activity, transferring acyl groups | | transferase activity, transferring acyl groups other than amino-acyl groups | | acyltransferase activity | | N-acyltransferase activity | | N-succinyltransferase activity | | 5-aminolevulinate synthase activity | | Process |
|---|
| heterocycle biosynthetic process | | tetrapyrrole biosynthetic process | | metabolic process | | biosynthetic process | | cellular biosynthetic process |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | chromosome 4 |
|---|
| Locus | YDR232W |
|---|
| Gene Sequence | >1647 bp
ATGCAACGCTCCATTTTTGCGAGGTTCGGTAACTCCTCTGCCGCTGTTTCCACACTGAAT
AGGCTGTCCACGACAGCCGCACCACATGCGAAAAATGGCTATGCCACCGCTACTGGTGCT
GGTGCCGCTGCTGCCACTGCCACAGCGTCATCAACACATGCAGCAGCAGCAGCAGCCGCT
GCTGCCAACCATTCCACCCAGGAGTCGGGTTTCGATTACGAAGGCCTGATAGATTCCGAA
CTGCAGAAGAAAAGACTTGACAAATCGTACAGATATTTCAACAATATCAACCGATTGGCC
AAGGAGTTCCCCCTAGCTCATCGCCAGAGAGAGGCGGACAAGGTCACCGTTTGGTGTTCC
AACGACTATTTAGCACTTTCCAAGCACCCTGAGGTATTGGACGCCATGCATAAAACTATC
GACAAGTATGGTTGTGGTGCCGGTGGTACAAGAAACATTGCTGGCCATAACATCCCCACT
TTGAATCTGGAAGCCGAATTGGCCACTTTACACAAGAAGGAAGGTGCCTTAGTTTTTTCG
TCATGTTACGTAGCCAACGATGCCGTCTTATCCCTACTGGGTCAAAAGATGAAGGACTTG
GTGATTTTCTCCGACGAACTCAACCATGCGTCCATGATTGTCGGTATTAAGCATGCTAAC
GTAAAAAAACACATTTTCAAACATAATGACTTGAACGAATTGGAACAACTGCTCCAGTCA
TACCCCAAATCCGTTCCTAAACTAATTGCTTTCGAATCAGTATATTCTATGGCCGGTTCA
GTGGCCGACATAGAAAAAATTTGCGACTTGGCCGACAAATACGGTGCTTTGACCTTCTTG
GATGAAGTACATGCGGTCGGCCTGTACGGCCCTCACGGTGCAGGTGTTGCAGAACATTGT
GATTTTGAAAGTCACCGTGCAAGTGGTATTGCTACCCCAAAGACCAATGACAAGGGCGGC
GCGAAGACTGTGATGGACCGTGTCGACATGATCACCGGCACTTTAGGTAAGTCTTTCGGT
AGCGTAGGTGGCTACGTCGCAGCCTCTAGGAAATTGATCGATTGGTTCAGATCGTTTGCA
CCTGGTTTCATTTTCACCACGACTTTACCACCTTCAGTTATGGCAGGCGCTACCGCAGCA
ATTAGATACCAACGTTGCCACATCGACCTAAGAACCTCGCAACAGAAACATACCATGTAC
GTAAAGAAAGCTTTCCATGAGTTGGGCATTCCAGTTATTCCAAATCCTTCTCATATCGTC
CCAGTGTTGATTGGTAATGCTGATTTGGCTAAGCAAGCTTCTGACATCTTAATCAATAAG
CATCAAATCTACGTACAAGCTATCAACTTCCCTACGGTTGCTCGCGGTACCGAAAGATTG
AGAATTACCCCAACGCCAGGTCACACCAACGATTTATCTGACATCTTAATCAATGCAGTT
GATGATGTGTTCAATGAGCTACAGTTACCACGTGTCAGAGACTGGGAAAGCCAAGGTGGC
TTATTGGGTGTTGGAGAGAGCGGATTTGTGGAAGAGTCTAACTTATGGACATCAAGCCAA
CTATCTTTAACTAATGACGACTTGAACCCTAATGTTAGAGACCCCATCGTTAAACAACTA
GAGGTTTCTAGTGGTATCAAGCAGTAA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | |
|---|
| Protein Residues | 548 |
|---|
| Protein Molecular Weight | 59361.69922 |
|---|
| Protein Theoretical pI | 7.56 |
|---|
| Signalling Regions | |
|---|
| Transmembrane Regions | |
|---|
| Protein Sequence | >5-aminolevulinate synthase, mitochondrial
MQRSIFARFGNSSAAVSTLNRLSTTAAPHAKNGYATATGAGAAAATATASSTHAAAAAAA
AANHSTQESGFDYEGLIDSELQKKRLDKSYRYFNNINRLAKEFPLAHRQREADKVTVWCS
NDYLALSKHPEVLDAMHKTIDKYGCGAGGTRNIAGHNIPTLNLEAELATLHKKEGALVFS
SCYVANDAVLSLLGQKMKDLVIFSDELNHASMIVGIKHANVKKHIFKHNDLNELEQLLQS
YPKSVPKLIAFESVYSMAGSVADIEKICDLADKYGALTFLDEVHAVGLYGPHGAGVAEHC
DFESHRASGIATPKTNDKGGAKTVMDRVDMITGTLGKSFGSVGGYVAASRKLIDWFRSFA
PGFIFTTTLPPSVMAGATAAIRYQRCHIDLRTSQQKHTMYVKKAFHELGIPVIPNPSHIV
PVLIGNADLAKQASDILINKHQIYVQAINFPTVARGTERLRITPTPGHTNDLSDILINAV
DDVFNELQLPRVRDWESQGGLLGVGESGFVEESNLWTSSQLSLTNDDLNPNVRDPIVKQL
EVSSGIKQ |
|---|
| References |
|---|
| External Links | |
|---|
| General Reference | - Urban-Grimal, D., Volland, C., Garnier, T., Dehoux, P., Labbe-Bois, R. (1986). "The nucleotide sequence of the HEM1 gene and evidence for a precursor form of the mitochondrial 5-aminolevulinate synthase in Saccharomyces cerevisiae." Eur J Biochem 156:511-519.3516694
- Jacq, C., Alt-Morbe, J., Andre, B., Arnold, W., Bahr, A., Ballesta, J. P., Bargues, M., Baron, L., Becker, A., Biteau, N., Blocker, H., Blugeon, C., Boskovic, J., Brandt, P., Bruckner, M., Buitrago, M. J., Coster, F., Delaveau, T., del Rey, F., Dujon, B., Eide, L. G., Garcia-Cantalejo, J. M., Goffeau, A., Gomez-Peris, A., Zaccaria, P., et, a. l. .. (1997). "The nucleotide sequence of Saccharomyces cerevisiae chromosome IV." Nature 387:75-78.9169867
- Keng, T., Guarente, L. (1987). "Constitutive expression of the yeast HEM1 gene is actually a composite of activation and repression." Proc Natl Acad Sci U S A 84:9113-9117.3321068
- Keng, T., Alani, E., Guarente, L. (1986). "The nine amino-terminal residues of delta-aminolevulinate synthase direct beta-galactosidase into the mitochondrial matrix." Mol Cell Biol 6:355-364.3023841
- Ghaemmaghami, S., Huh, W. K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., O'Shea, E. K., Weissman, J. S. (2003). "Global analysis of protein expression in yeast." Nature 425:737-741.14562106
|
|---|

