Acetylornithine aminotransferase, mitochondrial (P18544)
| Identification | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Acetylornithine aminotransferase, mitochondrial | ||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||
| Gene Name | ARG8 | ||||||||||||||||||||||||
| Enzyme Class | |||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||
| General Function | Involved in transaminase activity | ||||||||||||||||||||||||
| Specific Function | N(2)-acetyl-L-ornithine + 2-oxoglutarate = N- acetyl-L-glutamate 5-semialdehyde + L-glutamate | ||||||||||||||||||||||||
| Cellular Location | Mitochondrion matrix | ||||||||||||||||||||||||
| SMPDB Pathways |
| ||||||||||||||||||||||||
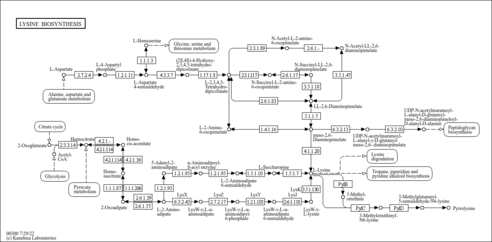
| KEGG Pathways |
| ||||||||||||||||||||||||
| SMPDB Reactions |
| ||||||||||||||||||||||||
| KEGG Reactions |
| ||||||||||||||||||||||||
| Metabolites |
| ||||||||||||||||||||||||
| GO Classification |
| ||||||||||||||||||||||||
| Gene Properties | |||||||||||||||||||||||||
| Chromosome Location | chromosome 15 | ||||||||||||||||||||||||
| Locus | YOL140W | ||||||||||||||||||||||||
| Gene Sequence | >1272 bp ATGTTTAAAAGATATTTATCCAGTACGTCATCAAGAAGATTTACAAGCATTTTAGAGGAA AAGGCCTTTCAAGTGACCACTTACTCTAGACCTGAAGATCTATGTATAACTAGAGGTAAA AATGCAAAGCTGTATGATGACGTGAATGGTAAAGAATATATCGATTTCACCGCAGGTATT GCGGTGACCGCATTAGGCCATGCAAATCCTAAAGTGGCAGAAATTCTGCACCATCAGGCT AACAAACTGGTTCATTCCTCCAACCTTTACTTCACTAAGGAATGTTTGGATTTAAGTGAA AAGATTGTTGAAAAGACCAAGCAATTCGGTGGTCAACACGACGCCTCAAGAGTATTTTTA TGTAATTCTGGTACGGAAGCAAATGAAGCTGCTTTGAAGTTTGCAAAGAAACATGGTATA ATGAAAAATCCTAGCAAGCAAGGCATTGTTGCATTTGAGAACTCTTTTCATGGCCGTACT ATGGGCGCTTTATCTGTCACTTGGAATAGTAAATATAGAACTCCTTTTGGGGATTTGGTT CCCCATGTCTCATTCTTAAATTTGAATGACGAAATGACCAAACTACAAAGTTATATCGAG ACCAAAAAGGACGAGATTGCTGGTTTAATTGTCGAGCCCATACAAGGTGAAGGTGGGGTT TTTCCCGTAGAAGTTGAAAAGCTAACCGGATTGAAGAAAATATGTCAAGATAATGATGTG ATTGTCATTCATGATGAAATTCAATGCGGTTTGGGCCGTTCAGGTAAACTATGGGCTCAT GCTTATTTACCAAGTGAGGCTCATCCGGATATTTTTACATCTGCCAAAGCATTGGGAAAT GGCTTCCCCATCGCTGCCACCATCGTCAATGAAAAAGTTAATAATGCTTTGAGAGTTGGT GACCACGGCACCACGTATGGTGGTAATCCGCTGGCCTGTTCTGTAAGCAACTATGTTTTG GATACCATAGCAGACGAAGCTTTTTTGAAACAAGTCTCTAAGAAGAGTGATATCTTACAA AAGCGCTTGCGCGAAATTCAAGCCAAATATCCAAATCAAATAAAGACTATCAGAGGAAAA GGTTTGATGCTTGGTGCTGAGTTCGTCGAACCACCCACCGAGGTCATCAAAAAGGCCAGA GAATTGGGACTTTTGATCATTACCGCTGGTAAGAGTACCGTTAGATTTGTTCCCGCATTA ACGATTGAAGACGAACTAATCGAAGAAGGGATGGATGCTTTTGAAAAGGCTATTGAAGCG GTTTACGCTTAA | ||||||||||||||||||||||||
| Protein Properties | |||||||||||||||||||||||||
| Pfam Domain Function |
| ||||||||||||||||||||||||
| Protein Residues | 423 | ||||||||||||||||||||||||
| Protein Molecular Weight | 46681.10156 | ||||||||||||||||||||||||
| Protein Theoretical pI | 7.62 | ||||||||||||||||||||||||
| Signalling Regions |
| ||||||||||||||||||||||||
| Transmembrane Regions |
| ||||||||||||||||||||||||
| Protein Sequence | >Acetylornithine aminotransferase, mitochondrial MFKRYLSSTSSRRFTSILEEKAFQVTTYSRPEDLCITRGKNAKLYDDVNGKEYIDFTAGI AVTALGHANPKVAEILHHQANKLVHSSNLYFTKECLDLSEKIVEKTKQFGGQHDASRVFL CNSGTEANEAALKFAKKHGIMKNPSKQGIVAFENSFHGRTMGALSVTWNSKYRTPFGDLV PHVSFLNLNDEMTKLQSYIETKKDEIAGLIVEPIQGEGGVFPVEVEKLTGLKKICQDNDV IVIHDEIQCGLGRSGKLWAHAYLPSEAHPDIFTSAKALGNGFPIAATIVNEKVNNALRVG DHGTTYGGNPLACSVSNYVLDTIADEAFLKQVSKKSDILQKRLREIQAKYPNQIKTIRGK GLMLGAEFVEPPTEVIKKARELGLLIITAGKSTVRFVPALTIEDELIEEGMDAFEKAIEA VYA | ||||||||||||||||||||||||
| References | |||||||||||||||||||||||||
| External Links |
| ||||||||||||||||||||||||
| General Reference |
| ||||||||||||||||||||||||