| Identification |
|---|
| Name | Mevalonate kinase |
|---|
| Synonyms | - MK
- MvK
- Ergosterol biosynthesis protein 12
- Regulation of autonomous replication protein 1
|
|---|
| Gene Name | ERG12 |
|---|
| Enzyme Class | |
|---|
| Biological Properties |
|---|
| General Function | Involved in ATP binding |
|---|
| Specific Function | RAR (regulation of autonomous replication) is a protein whose activity increases the mitotic stability of plasmids |
|---|
| Cellular Location | Cytoplasm |
|---|
| SMPDB Pathways | | Cholesterol biosynthesis and metabolism CE(10:0) | PW002545 | 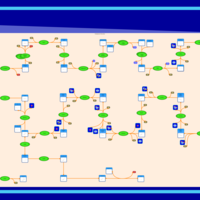 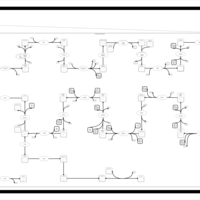 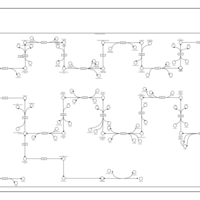 | | Cholesterol biosynthesis and metabolism CE(12:0) | PW002548 | 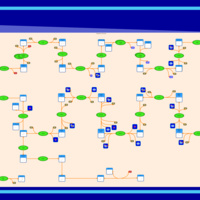 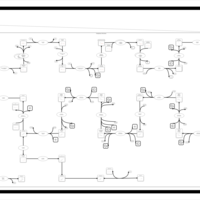 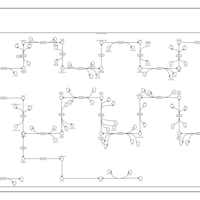 | | Cholesterol biosynthesis and metabolism CE(14:0) | PW002544 | 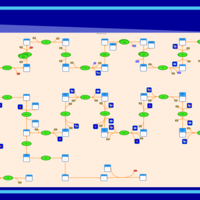 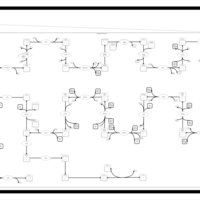 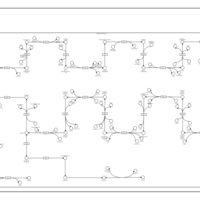 | | Cholesterol biosynthesis and metabolism CE(16:0) | PW002550 | 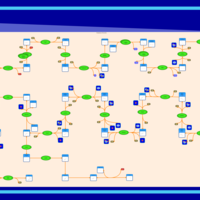 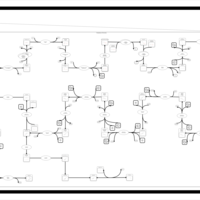 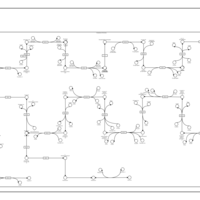 | | Cholesterol biosynthesis and metabolism CE(18:0) | PW002551 |    |
|
|---|
| KEGG Pathways | | Terpenoid backbone biosynthesis | ec00900 | 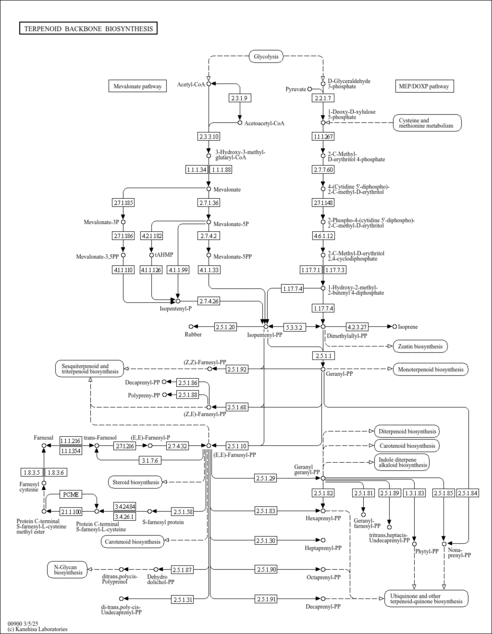 |
|
|---|
| SMPDB Reactions | |
|---|
| KEGG Reactions | |
|---|
| Metabolites | | YMDB ID | Name | View |
|---|
| YMDB00064 | CDP | Show | | YMDB00109 | Adenosine triphosphate | Show | | YMDB00181 | Mevalonic acid-5P | Show | | YMDB00257 | Guanosine diphosphate | Show | | YMDB00279 | Cytidine triphosphate | Show | | YMDB00307 | Uridine 5'-diphosphate | Show | | YMDB00310 | 22b-Hydroxycholesterol | Show | | YMDB00326 | Uridine triphosphate | Show | | YMDB00558 | GTP | Show | | YMDB00707 | (R)-Mevalonic acid | Show | | YMDB00862 | hydron | Show | | YMDB00914 | ADP | Show | | YMDB01039 | (R)-Mevalonate | Show |
|
|---|
| GO Classification | | Component |
|---|
| intracellular part | | cytoplasm | | cell part | | Function |
|---|
| binding | | mevalonate kinase activity | | nucleoside binding | | purine nucleoside binding | | adenyl nucleotide binding | | adenyl ribonucleotide binding | | ATP binding | | kinase activity | | catalytic activity | | phosphotransferase activity, alcohol group as acceptor | | transferase activity | | transferase activity, transferring phosphorus-containing groups | | Process |
|---|
| isoprenoid metabolic process | | isoprenoid biosynthetic process | | primary metabolic process | | phosphorus metabolic process | | metabolic process | | phosphate metabolic process | | phosphorylation | | lipid metabolic process | | cellular lipid metabolic process | | cellular metabolic process |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | chromosome 13 |
|---|
| Locus | YMR208W |
|---|
| Gene Sequence | >1332 bp
ATGTCATTACCGTTCTTAACTTCTGCACCGGGAAAGGTTATTATTTTTGGTGAACACTCT
GCTGTGTACAACAAGCCTGCCGTCGCTGCTAGTGTGTCTGCGTTGAGAACCTACCTGCTA
ATAAGCGAGTCATCTGCACCAGATACTATTGAATTGGACTTCCCGGACATTAGCTTTAAT
CATAAGTGGTCCATCAATGATTTCAATGCCATCACCGAGGATCAAGTAAACTCCCAAAAA
TTGGCCAAGGCTCAACAAGCCACCGATGGCTTGTCTCAGGAACTCGTTAGTCTTTTGGAT
CCGTTGTTAGCTCAACTATCCGAATCCTTCCACTACCATGCAGCGTTTTGTTTCCTGTAT
ATGTTTGTTTGCCTATGCCCCCATGCCAAGAATATTAAGTTTTCTTTAAAGTCTACTTTA
CCCATCGGTGCTGGGTTGGGCTCAAGCGCCTCTATTTCTGTATCACTGGCCTTAGCTATG
GCCTACTTGGGGGGGTTAATAGGATCTAATGACTTGGAAAAGCTGTCAGAAAACGATAAG
CATATAGTGAATCAATGGGCCTTCATAGGTGAAAAGTGTATTCACGGTACCCCTTCAGGA
ATAGATAACGCTGTGGCCACTTATGGTAATGCCCTGCTATTTGAAAAAGACTCACATAAT
GGAACAATAAACACAAACAATTTTAAGTTCTTAGATGATTTCCCAGCCATTCCAATGATC
CTAACCTATACTAGAATTCCAAGGTCTACAAAAGATCTTGTTGCTCGCGTTCGTGTGTTG
GTCACCGAGAAATTTCCTGAAGTTATGAAGCCAATTCTAGATGCCATGGGTGAATGTGCC
CTACAAGGCTTAGAGATCATGACTAAGTTAAGTAAATGTAAAGGCACCGATGACGAGGCT
GTAGAAACTAATAATGAACTGTATGAACAACTATTGGAATTGATAAGAATAAATCATGGA
CTGCTTGTCTCAATCGGTGTTTCTCATCCTGGATTAGAACTTATTAAAAATCTGAGCGAT
GATTTGAGAATTGGCTCCACAAAACTTACCGGTGCTGGTGGCGGCGGTTGCTCTTTGACT
TTGTTACGAAGAGACATTACTCAAGAGCAAATTGACAGCTTCAAAAAGAAATTGCAAGAT
GATTTTAGTTACGAGACATTTGAAACAGACTTGGGTGGGACTGGCTGCTGTTTGTTAAGC
GCAAAAAATTTGAATAAAGATCTTAAAATCAAATCCCTAGTATTCCAATTATTTGAAAAT
AAAACTACCACAAAGCAACAAATTGACGATCTATTATTGCCAGGAAACACGAATTTACCA
TGGACTTCATAA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | |
|---|
| Protein Residues | 443 |
|---|
| Protein Molecular Weight | 48458.89844 |
|---|
| Protein Theoretical pI | 5.28 |
|---|
| Signalling Regions | |
|---|
| Transmembrane Regions | |
|---|
| Protein Sequence | >Mevalonate kinase
MSLPFLTSAPGKVIIFGEHSAVYNKPAVAASVSALRTYLLISESSAPDTIELDFPDISFN
HKWSINDFNAITEDQVNSQKLAKAQQATDGLSQELVSLLDPLLAQLSESFHYHAAFCFLY
MFVCLCPHAKNIKFSLKSTLPIGAGLGSSASISVSLALAMAYLGGLIGSNDLEKLSENDK
HIVNQWAFIGEKCIHGTPSGIDNAVATYGNALLFEKDSHNGTINTNNFKFLDDFPAIPMI
LTYTRIPRSTKDLVARVRVLVTEKFPEVMKPILDAMGECALQGLEIMTKLSKCKGTDDEA
VETNNELYEQLLELIRINHGLLVSIGVSHPGLELIKNLSDDLRIGSTKLTGAGGGGCSLT
LLRRDITQEQIDSFKKKLQDDFSYETFETDLGGTGCCLLSAKNLNKDLKIKSLVFQLFEN
KTTTKQQIDDLLLPGNTNLPWTS |
|---|
| References |
|---|
| External Links | |
|---|
| General Reference | - Kearsey, S. E., Edwards, J. (1987). "Mutations that increase the mitotic stability of minichromosomes in yeast: characterization of RAR1." Mol Gen Genet 210:509-517.3323847
- Oulmouden, A., Karst, F. (1991). "Nucleotide sequence of the ERG12 gene of Saccharomyces cerevisiae encoding mevalonate kinase." Curr Genet 19:9-14.1645230
- Bowman, S., Churcher, C., Badcock, K., Brown, D., Chillingworth, T., Connor, R., Dedman, K., Devlin, K., Gentles, S., Hamlin, N., Hunt, S., Jagels, K., Lye, G., Moule, S., Odell, C., Pearson, D., Rajandream, M., Rice, P., Skelton, J., Walsh, S., Whitehead, S., Barrell, B. (1997). "The nucleotide sequence of Saccharomyces cerevisiae chromosome XIII." Nature 387:90-93.9169872
- Ghaemmaghami, S., Huh, W. K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., O'Shea, E. K., Weissman, J. S. (2003). "Global analysis of protein expression in yeast." Nature 425:737-741.14562106
|
|---|
