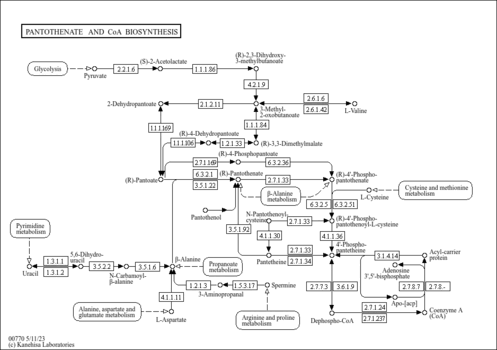
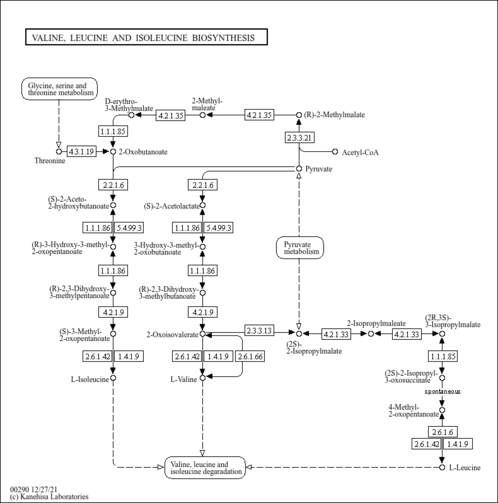
| General Reference | - Kispal, G., Steiner, H., Court, D. A., Rolinski, B., Lill, R. (1996). "Mitochondrial and cytosolic branched-chain amino acid transaminases from yeast, homologs of the myc oncogene-regulated Eca39 protein." J Biol Chem 271:24458-24464.8798704
- Johnston, M., Andrews, S., Brinkman, R., Cooper, J., Ding, H., Dover, J., Du, Z., Favello, A., Fulton, L., Gattung, S., et, a. l. .. (1994). "Complete nucleotide sequence of Saccharomyces cerevisiae chromosome VIII." Science 265:2077-2082.8091229
- Hu, Y., Rolfs, A., Bhullar, B., Murthy, T. V., Zhu, C., Berger, M. F., Camargo, A. A., Kelley, F., McCarron, S., Jepson, D., Richardson, A., Raphael, J., Moreira, D., Taycher, E., Zuo, D., Mohr, S., Kane, M. F., Williamson, J., Simpson, A., Bulyk, M. L., Harlow, E., Marsischky, G., Kolodner, R. D., LaBaer, J. (2007). "Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae." Genome Res 17:536-543.17322287
- Eden, A., Simchen, G., Benvenisty, N. (1996). "Two yeast homologs of ECA39, a target for c-Myc regulation, code for cytosolic and mitochondrial branched-chain amino acid aminotransferases." J Biol Chem 271:20242-20245.8702755
- Ben-Yosef, T., Yanuka, O., Benvenisty, N. (1996). "ECA39 is regulated by c-Myc in human and by a Jun/Fos homolog, Gcn4, in yeast." Oncogene 13:1859-1866.8934531
- Ghaemmaghami, S., Huh, W. K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., O'Shea, E. K., Weissman, J. S. (2003). "Global analysis of protein expression in yeast." Nature 425:737-741.14562106
- Albuquerque, C. P., Smolka, M. B., Payne, S. H., Bafna, V., Eng, J., Zhou, H. (2008). "A multidimensional chromatography technology for in-depth phosphoproteome analysis." Mol Cell Proteomics 7:1389-1396.18407956
|
|---|