| Identification |
|---|
| Name | Probable phospholipid-transporting ATPase DNF3 |
|---|
| Synonyms | Not Available |
|---|
| Gene Name | DNF3 |
|---|
| Enzyme Class | |
|---|
| Biological Properties |
|---|
| General Function | Involved in nucleotide binding |
|---|
| Specific Function | This magnesium-dependent enzyme catalyzes the hydrolysis of ATP coupled with the transport of phospholipids (Potential) |
|---|
| Cellular Location | Membrane; Multi-pass membrane protein |
|---|
| SMPDB Pathways | | Lysolipid incorporation into ER | PW002532 | 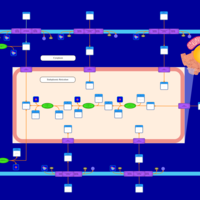 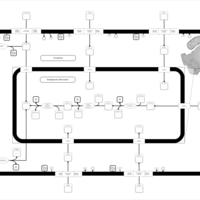 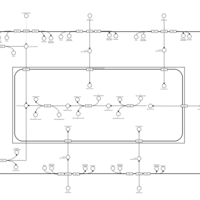 | | Lysolipid incorporation into ER PC(10:0/10:0) | PW002780 | 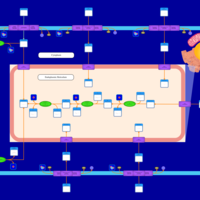 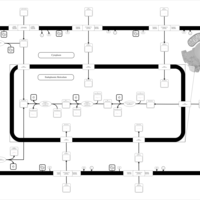 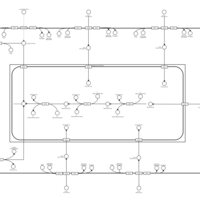 | | Lysolipid incorporation into ER PC(14:0/14:0) | PW002783 | 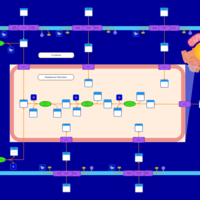 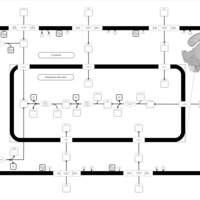 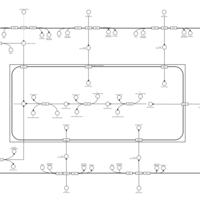 | | Lysolipid incorporation into ER PC(16:0/16:0) | PW002784 | 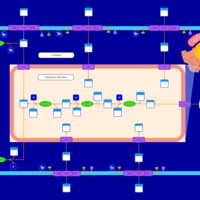 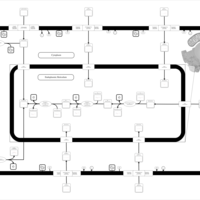 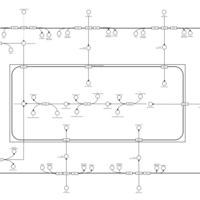 | | Lysolipid incorporation into ER PC(16:1(11Z)/16:1(11Z)) | PW002786 | 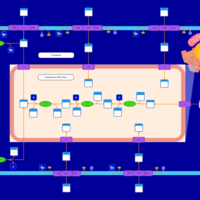 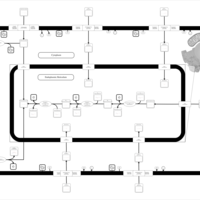 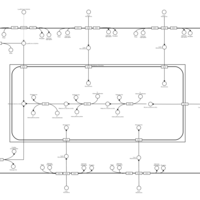 |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Metabolites | | YMDB ID | Name | View |
|---|
| YMDB00109 | Adenosine triphosphate | Show | | YMDB00890 | water | Show | | YMDB00907 | phosphate | Show | | YMDB00914 | ADP | Show |
|
|---|
| GO Classification | | Component |
|---|
| membrane part | | intrinsic to membrane | | integral to membrane | | cell part | | membrane | | Function |
|---|
| ATP binding | | ion binding | | cation binding | | substrate-specific transporter activity | | metal ion binding | | lipid transporter activity | | transporter activity | | phospholipid transporter activity | | transmembrane transporter activity | | phospholipid-translocating ATPase activity | | hydrolase activity | | substrate-specific transmembrane transporter activity | | ion transmembrane transporter activity | | magnesium ion binding | | binding | | ATPase activity, coupled to transmembrane movement of ions | | catalytic activity | | nucleoside binding | | ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism | | purine nucleoside binding | | hydrolase activity, acting on acid anhydrides | | adenyl nucleotide binding | | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances | | adenyl ribonucleotide binding | | Process |
|---|
| purine nucleoside triphosphate biosynthetic process | | purine ribonucleoside triphosphate biosynthetic process | | ATP biosynthetic process | | nitrogen compound metabolic process | | cellular nitrogen compound metabolic process | | nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | | nucleobase, nucleoside and nucleotide metabolic process | | establishment of localization | | transport | | lipid transport | | nucleoside phosphate metabolic process | | phospholipid transport | | nucleotide metabolic process | | purine nucleotide metabolic process | | purine nucleotide biosynthetic process | | metabolic process |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | chromosome 13 |
|---|
| Locus | YMR162C |
|---|
| Gene Sequence | >4971 bp
ATGGGCATAGCTGATGGGCAAAGAAGAAGATCTTCATCACTACGAACGCAGATGTTCAAC
AAGCATTTGTATGATAAATATCGCGGAAGAACTGATGATGAGATAGAGTTAGAAGATATA
AATGAGAGTAAGACGTTCTCCGGTAGCGATAATAACGATAAAGATGACAGAGATGAAACA
AGCGGCAATTATGCTGCTGAAGAGGATTATGAAATGGAAGAGTATGGTAGTCCTGATGTG
TCATATAGCATTATCACTAAAATACTTGATACTATCCTAGATCGAAGGAGGACGTTCCAT
TCGAAAGACGGTAGACATATTCCGATTATATTAGATCATAATGCAATTGAATACAAACAA
GCTGCTACTAAAAGGGATGGTCATTTGATAGACGAAAGGTTCAACAAGCCTTACTGCGAC
AACCGAATAACTTCATCAAGATATACATTCTATTCCTTTTTGCCTAGACAGTTGTATGCA
CAATTTTCAAAGTTAGCCAATACTTATTTTTTCATTGTCGCAGTATTACAAATGATACCT
GGATGGTCTACAACTGGTACTTATACAACTATTATCCCCTTATGTGTATTTATGGGTATA
TCCATGACAAGAGAGGCTTGGGACGATTTCAGGAGACATCGTTTAGATAAGGAAGAGAAC
AATAAACCAGTGGGAGTGTTGGTTAAAGATGGAAATAATGACGCACAAGAAGTTTATACA
CTACCAAGTAGTGTTGTCTCATCCACTGCATATCTAACTAAATCGGCCGCTGCTGAAAAT
AATCCACCTTTAAACGATGATCGGAACTCCTCTCAAGGACACTTCTTAGACACGCACTTC
AATAACTTTGAACTACTCAAGAATAAATACAATGTTCATATTCATCAAAAGAAGTGGGAA
AAACTTCGTGTTGGAGACTTCGTACTTCTCACACAAGATGATTGGGTACCTGCAGATCTG
CTTTTATTGACATGTGACGGCGAAAATAGCGAATGTTTCGTTGAAACGATGGCTTTAGAT
GGCGAAACAAATTTGAAAAGTAAACAACCCCATCCAGAATTAAATAAGTTAACCAAAGCC
GCATCAGGTCTTGCAAATATTAATGCTCAAGTTACCGTGGAAGATCCCAATATTGATTTA
TATAATTTTGAAGGTAATCTTGAATTGAAAAACCATCGCAATGATACCATAATGAAATAC
CCTCTAGGTCCTGATAATGTCATTTATCGTGGTAGTATTTTGAGGAATACTCAAAATGTT
GTTGGTATGGTCATTTTCAGTGGTGAGGAAACAAAAATCAGAATGAATGCTTTAAAAAAC
CCAAGAACGAAAGCCCCTAAGCTACAAAGGAAGATAAATATGATTATCGTGTTTATGGTG
TTTGTCGTTGCCACAATATCTTTATTTTCTTATCTTGGCCATGTTTTGCATAAGAAAAAA
TATATCGACCAGAACAAAGCATGGTATCTATTTCAGGCAGATGCTGGCGTAGCACCCACA
ATAATGTCTTTTATCATCATGTATAACACTGTGATTCCTTTGTCCCTATATGTAACAATG
GAAATTATCAAGGTAGTCCAAAGTAAAATGATGGAATGGGACATAGACATGTACCATGCA
GAAACGAACACGCCATGCGAATCGAGAACTGCCACCATTCTGGAGGAATTGGGGCAAGTA
TCTTATATTTTTAGTGATAAAACCGGAACGCTGACGGATAATAAGATGATATTTCGGAAA
TTTTCTTTATGTGGATCATCTTGGCTGCATAATGTAGATCTCGGTAATTCTGAGGATAAC
TTTGAAGACAACAGAGACAATACGAACTCCTTGAGACTACCACCAAAAGCTCACAACGGC
TCTAGTATAGACGTTGTTTCCATTGGAGATCAAAATGTTCTTGATCGGTTAGGATTTTCG
GACGCACCAATTGAAAAAGGGCATCGTCCATCTTTGGACAACTTTCCTAAATCTAGGAAT
TCTATTGAATATAAGGGAAATTCATCAGCAATTTATACAGGCCGTCCCAGTATGAGATCA
CTTTTTGGGAAAGATAATTCACATCTAAGTAAACAAGCTTCGGTTATTTCACCCTCCGAG
ACATTTTCGGAGAACATTAAAAGTTCGTTTGATTTAATTCAGTTTATTCAAAGATATCCG
ACCGCCCTGTTTTCTCAAAAGGCTAAATTTTTCTTCTTATCATTAGCTCTTTGTCATAGC
TGTCTTCCCAAAAAAACTCATAACGAATCAATAGGAGAAGACTCCATAGAGTATCAGTCT
TCATCTCCAGATGAATTAGCACTAGTAACCGCTGCTAGAGACCTTGGTTATATTGTATTG
AATAGAAATGCACAGATTTTGACCATTAAAACTTTTCCAGACGGTTTTGACGGTGAAGCA
AAATTGGAAAATTATGAAATTTTGAACTATATTGACTTCAATTCTCAAAGAAAGAGAATG
TCTGTTCTTGTACGTATGCCAAATCAACCAAATCAAGTTCTTTTAATTTGCAAAGGTGCT
GATAACGTTATTATGGAGCGTTTACATGATCGCGAATTGGCTGCCAAAAAAATGGCAGAC
ATATGCACCAGTACCAAAGAACGTAAAGACGCAGAAGCAGAGCTGGTTCTCCAGCAAAGA
AAGTCTCTGGAAAGAATGGTAGATGAAGAGGCTATGGCCAGGACCTCATTGAGAAATTCT
TTGTCTAGCGTACCGAGAGCAAGTTTATCATTGCAGGCAGTGAGAAAAAGTCTTTCAATG
AAGAACAGCAGGACCCGTGATCCAGAGAAACAAATTGACTCCATTGATCAATTTCTAGAA
ACGGTCAAGAAATCTGATCAGGAGATTGGCAGCGTTGTAAATAAAAGCAGGAAGTCACTC
CATAAGCAACAAATAGAAAAATATGGGCCGCGAATTTCGATAGATGGAACACATTTTCCG
AACAACAATGTCCCAATAGACACAAGAAAAGAAGGACTACAGCATGATTACGATACAGAA
ATTTTGGAGCACATTGGAAGCGATGAGTTAATTTTGAATGAAGAGTATGTTATTGAAAGA
ACTTTGCAAGCCATCGATGAGTTTTCGACTGAAGGCTTAAGAACTCTGGTCTATGCTTAC
AAATGGATCGATATTGGGCAGTATGAAAATTGGAATAAACGCTATCACCAAGCCAAAACC
TCCCTTACTGATAGAAAAATCAAAGTCGATGAGGCAGGTGCAGAAATCGAGGATGGTTTA
AATCTGTTAGGGGTAACTGCAATCGAAGACAAGTTACAAGACGGCGTTTCAGAAGCTATA
GAAAAGATCAGAAGAGCCGGTATAAAAATGTGGATGCTAACAGGGGATAAAAGGGAAACG
GCTATAAATATAGGGTATTCGTGTATGTTAATCAAAGATTATTCTACGGTAGTCATTTTA
ACTACTACTGATGAAAATATTATATCAAAGATGAATGCGGTTTCTCAAGAAGTTGATTCC
GGAAACATAGCACACTGCGTTGTTGTAATTGACGGTGCCACGATGGCCATGTTTGAAGGA
AATCCTACATACATGTCTGTATTTGTTGAGCTTTGCACTAAAACAGACTCGGTAATATGT
TGCCGTGCTTCACCTTCTCAAAAAGCCCTTATGGTTTCCAACATAAGAAATACCGATCCA
AACTTAGTCACGTTGGCAATAGGTGATGGTGCGAATGATATTGCCATGATACAATCTGCC
GATATAGGTGTTGGAATAGCCGGTAAGGAGGGATTGCAAGCATCTAGAGTATCTGATTAT
TCTATTGGCCAATTTCGGTTTCTCTTGAAATTGTTATTTGTTCATGGGCGTTATAACTAT
ATTCGCACCTCCAAGTTTATGCTTTGCACGTTTTACAAAGAAATTACGTTTTACTTTACG
CAGTTGATATATCAAAGATATACAATGTTTTCAGGCTCCTCTCTATATGAGCCATGGTCA
TTGTCCATGTTCAATACTCTATTCACATCACTACCGGTTCTATGTATTGGTATGTTTGAG
AAAGACTTGAAACCAATGACTCTACTAACAGTCCCTGAATTATATTCGTACGGGCGCCTT
TCACAAGGTTTTAATTGGTTAATATTCATGGAGTGGGTTATCTTGGCAACTACGAATTCT
CTTATCATTACATTTTTAAATGTTGTAATGTGGGGTATGAGTTCTCTTTCAGATAACACT
ATGTATCCTCTAGGTCTCATCAATTTCACAGCCATTGTTGCCTTAATTAACGTTAAATCC
CAATTTGTTGAAATGCACAATAGGAATTGGCTGGCTTTTACTTCTGTAGTTCTATCCTGT
GGAGGTTGGTTAGTGTGGTGTTGTGCCCTTCCTATTTTGAATAATACTGACCAAATATAT
GATGTAGCATATGGCTTTTACAATCATTTTGGTAAAGACATTACATTCTGGTGTACTAGC
TTGGTGTTGGCACTGTTACCTATCACTTTAGATATAGTTTACAAAACATTCAAAGTTATG
ATATGGCCATCAGATTCTGATATTTTTGCTGAGCTGGAACAAAAAAGTGATATCAGAAAG
AAGCTGGAACTAGGAGCCTATAGCGAAATGAGGCAAGGATGGACGTGGGATAAAGATCCC
AGTACTTTCACAAGGTACACAGACAAAGTTCTTTCAAGGCCAAGAACTAATTCACGTGCA
AGTGCAAAAACTCATAACAGCAGCATATATTCAATGTCAAATGGGAATGTAGACCATTCC
TCAAAAAAAAATTTCTTTGGAAACTCTTCTAAAAAGAGCTCTGAACGATACGAGGTATTA
CCTAGTGGCAAGTTAATCAAGCGACCATCCTTGAAAACACAAAGCAGTAAAGACAGTATT
GGCGGCAATATAACTACTAAACTGACGAAGAAACTGAAGCTTCCATCGAGGAATGTGGAA
GATGAAGATGTGAACCAGATTATTCAGGCAAGGCTAAAGGATCTGGAGTAA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | |
|---|
| Protein Residues | 1656 |
|---|
| Protein Molecular Weight | 188318.0 |
|---|
| Protein Theoretical pI | 6.74 |
|---|
| Signalling Regions | |
|---|
| Transmembrane Regions | - 165-185
- 452-472
- 496-516
- 1158-1178
- 1319-1339
- 1366-1386
- 1396-1416
- 1433-1453
- 1474-1494
|
|---|
| Protein Sequence | >Probable phospholipid-transporting ATPase DNF3
MGIADGQRRRSSSLRTQMFNKHLYDKYRGRTDDEIELEDINESKTFSGSDNNDKDDRDET
SGNYAAEEDYEMEEYGSPDVSYSIITKILDTILDRRRTFHSKDGRHIPIILDHNAIEYKQ
AATKRDGHLIDERFNKPYCDNRITSSRYTFYSFLPRQLYAQFSKLANTYFFIVAVLQMIP
GWSTTGTYTTIIPLCVFMGISMTREAWDDFRRHRLDKEENNKPVGVLVKDGNNDAQEVYT
LPSSVVSSTAYLTKSAAAENNPPLNDDRNSSQGHFLDTHFNNFELLKNKYNVHIHQKKWE
KLRVGDFVLLTQDDWVPADLLLLTCDGENSECFVETMALDGETNLKSKQPHPELNKLTKA
ASGLANINAQVTVEDPNIDLYNFEGNLELKNHRNDTIMKYPLGPDNVIYRGSILRNTQNV
VGMVIFSGEETKIRMNALKNPRTKAPKLQRKINMIIVFMVFVVATISLFSYLGHVLHKKK
YIDQNKAWYLFQADAGVAPTIMSFIIMYNTVIPLSLYVTMEIIKVVQSKMMEWDIDMYHA
ETNTPCESRTATILEELGQVSYIFSDKTGTLTDNKMIFRKFSLCGSSWLHNVDLGNSEDN
FEDNRDNTNSLRLPPKAHNGSSIDVVSIGDQNVLDRLGFSDAPIEKGHRPSLDNFPKSRN
SIEYKGNSSAIYTGRPSMRSLFGKDNSHLSKQASVISPSETFSENIKSSFDLIQFIQRYP
TALFSQKAKFFFLSLALCHSCLPKKTHNESIGEDSIEYQSSSPDELALVTAARDLGYIVL
NRNAQILTIKTFPDGFDGEAKLENYEILNYIDFNSQRKRMSVLVRMPNQPNQVLLICKGA
DNVIMERLHDRELAAKKMADICTSTKERKDAEAELVLQQRKSLERMVDEEAMARTSLRNS
LSSVPRASLSLQAVRKSLSMKNSRTRDPEKQIDSIDQFLETVKKSDQEIGSVVNKSRKSL
HKQQIEKYGPRISIDGTHFPNNNVPIDTRKEGLQHDYDTEILEHIGSDELILNEEYVIER
TLQAIDEFSTEGLRTLVYAYKWIDIGQYENWNKRYHQAKTSLTDRKIKVDEAGAEIEDGL
NLLGVTAIEDKLQDGVSEAIEKIRRAGIKMWMLTGDKRETAINIGYSCMLIKDYSTVVIL
TTTDENIISKMNAVSQEVDSGNIAHCVVVIDGATMAMFEGNPTYMSVFVELCTKTDSVIC
CRASPSQKALMVSNIRNTDPNLVTLAIGDGANDIAMIQSADIGVGIAGKEGLQASRVSDY
SIGQFRFLLKLLFVHGRYNYIRTSKFMLCTFYKEITFYFTQLIYQRYTMFSGSSLYEPWS
LSMFNTLFTSLPVLCIGMFEKDLKPMTLLTVPELYSYGRLSQGFNWLIFMEWVILATTNS
LIITFLNVVMWGMSSLSDNTMYPLGLINFTAIVALINVKSQFVEMHNRNWLAFTSVVLSC
GGWLVWCCALPILNNTDQIYDVAYGFYNHFGKDITFWCTSLVLALLPITLDIVYKTFKVM
IWPSDSDIFAELEQKSDIRKKLELGAYSEMRQGWTWDKDPSTFTRYTDKVLSRPRTNSRA
SAKTHNSSIYSMSNGNVDHSSKKNFFGNSSKKSSERYEVLPSGKLIKRPSLKTQSSKDSI
GGNITTKLTKKLKLPSRNVEDEDVNQIIQARLKDLE |
|---|
| References |
|---|
| External Links | |
|---|
| General Reference | - Bowman, S., Churcher, C., Badcock, K., Brown, D., Chillingworth, T., Connor, R., Dedman, K., Devlin, K., Gentles, S., Hamlin, N., Hunt, S., Jagels, K., Lye, G., Moule, S., Odell, C., Pearson, D., Rajandream, M., Rice, P., Skelton, J., Walsh, S., Whitehead, S., Barrell, B. (1997). "The nucleotide sequence of Saccharomyces cerevisiae chromosome XIII." Nature 387:90-93.9169872
- Kim, H., Melen, K., Osterberg, M., von Heijne, G. (2006). "A global topology map of the Saccharomyces cerevisiae membrane proteome." Proc Natl Acad Sci U S A 103:11142-11147.16847258
- Chi, A., Huttenhower, C., Geer, L. Y., Coon, J. J., Syka, J. E., Bai, D. L., Shabanowitz, J., Burke, D. J., Troyanskaya, O. G., Hunt, D. F. (2007). "Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry." Proc Natl Acad Sci U S A 104:2193-2198.17287358
- Albuquerque, C. P., Smolka, M. B., Payne, S. H., Bafna, V., Eng, J., Zhou, H. (2008). "A multidimensional chromatography technology for in-depth phosphoproteome analysis." Mol Cell Proteomics 7:1389-1396.18407956
|
|---|