| Identification |
|---|
| Name | Serine hydroxymethyltransferase, cytosolic |
|---|
| Synonyms | - SHMT
- Glycine hydroxymethyltransferase
- Serine methylase
|
|---|
| Gene Name | SHM2 |
|---|
| Enzyme Class | |
|---|
| Biological Properties |
|---|
| General Function | Involved in catalytic activity |
|---|
| Specific Function | Interconversion of serine and glycine |
|---|
| Cellular Location | Cytoplasm |
|---|
| SMPDB Pathways | |
|---|
| KEGG Pathways | | Cyanoamino acid metabolism | ec00460 | 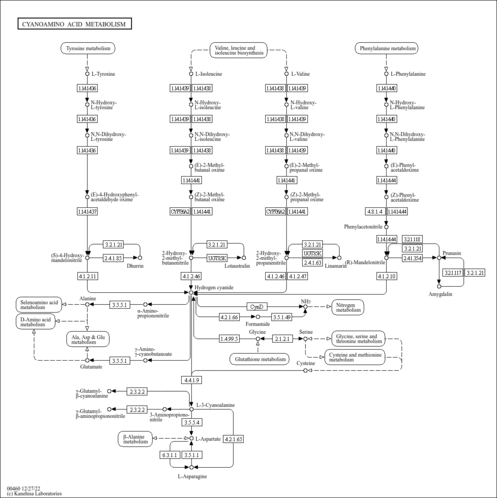 | | Glycine, serine and threonine metabolism | ec00260 |  | | Methane metabolism | ec00680 | 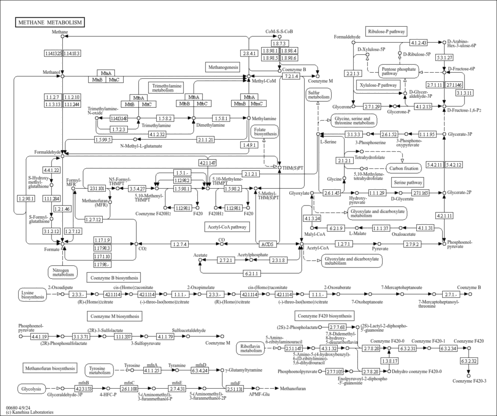 | | One carbon pool by folate | ec00670 | 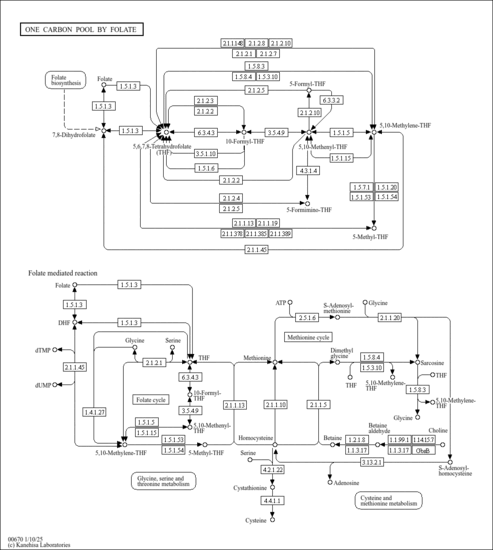 |
|
|---|
| SMPDB Reactions | |
|---|
| KEGG Reactions | |
|---|
| Metabolites | | YMDB ID | Name | View |
|---|
| YMDB00016 | Glycine | Show | | YMDB00112 | L-Serine | Show | | YMDB00139 | Tetrahydrofolic acid | Show | | YMDB00260 | 5,10-Methylene-THF | Show | | YMDB00483 | 5,6,7,8-Tetrahydrofolic acid | Show | | YMDB00848 | (6R)-5,10-methylenetetrahydrofolic acid | Show | | YMDB00890 | water | Show |
|
|---|
| GO Classification | | Component |
|---|
| Not Available | | Function |
|---|
| binding | | cofactor binding | | pyridoxal phosphate binding | | catalytic activity | | transferase activity | | transferase activity, transferring one-carbon groups | | methyltransferase activity | | glycine hydroxymethyltransferase activity | | Process |
|---|
| cellular amino acid metabolic process | | serine family amino acid metabolic process | | glycine metabolic process | | L-serine metabolic process | | metabolic process | | cellular metabolic process | | cellular amino acid and derivative metabolic process |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | chromosome 12 |
|---|
| Locus | YLR058C |
|---|
| Gene Sequence | >1410 bp
ATGCCTTACACTCTATCCGACGCTCATCATAAGTTGATCACCTCTCATTTGGTGGACACC
GACCCTGAAGTGGACTCCATTATCAAGGATGAAATTGAAAGACAAAAGCACTCCATCGAT
TTGATTGCTTCTGAAAATTTCACCTCAACCTCCGTTTTCGATGCCCTTGGAACTCCATTG
TCCAACAAATATTCTGAAGGTTATCCAGGTGCTCGTTACTACGGTGGTAATGAACACATT
GACAGAATGGAAATTCTATGTCAACAAAGAGCTTTGAAAGCTTTCCATGTTACTCCAGAC
AAATGGGGTGTTAACGTCCAAACTTTATCTGGTTCTCCTGCTAACTTGCAAGTTTATCAA
GCTATTATGAAGCCTCATGAAAGATTGATGGGTCTATACCTACCAGATGGTGGTCATTTG
TCTCACGGTTACGCTACTGAAAACAGAAAAATTTCTGCTGTTTCCACATACTTTGAATCT
TTCCCATACAGAGTTAACCCAGAAACCGGTATTATCGACTACGATACTTTAGAAAAGAAC
GCCATCCTATATAGACCAAAGGTTCTTGTTGCTGGTACTTCAGCATACTGTCGTTTAATT
GACTACAAGAGAATGAGAGAAATCGCCGACAAATGTGGTGCTTACTTGATGGTAGACATG
GCCCACATTTCTGGTTTGATCGCCGCAGGTGTCATCCCATCTCCTTTCGAATACGCTGAT
ATCGTTACCACCACCACTCACAAGTCTTTGAGAGGTCCACGTGGTGCTATGATTTTCTTC
AGAAGAGGTGTGAGATCTATCAACCCTAAGACCGGTAAGGAAGTTCTATACGACTTGGAA
AACCCAATTAACTTCTCTGTTTTCCCAGGTCACCAAGGTGGTCCACACAACCATACCATT
GCTGCTTTGGCCACTGCTTTGAAGCAAGCTGCCACTCCAGAATTCAAGGAATACCAAACT
CAAGTCTTGAAGAATGCTAAGGCTTTGGAAAGTGAATTTAAGAACTTGGGCTACAGATTA
GTTTCCAACGGTACCGATTCTCACATGGTTCTAGTATCCTTGAGAGAAAAGGGTGTTGAT
GGTGCTCGTGTTGAATACATTTGTGAAAAGATTAACATTGCTTTGAACAAAAATTCTATT
CCAGGTGACAAATCTGCTTTGGTTCCAGGTGGTGTCCGTATTGGGGCTCCAGCCATGACC
ACTAGAGGAATGGGTGAAGAAGATTTCCACAGAATTGTTCAATACATTAACAAGGCTGTA
GAATTCGCTCAACAAGTTCAACAAAAGCTTCCAAAGGATGCTTGTAGATTAAAGGACTTC
AAAGCCAAGGTCGACGAAGGCTCTGATGTTTTGAACACCTGGAAAAAGGAAATTTACGAC
TGGGCTGGCGAATACCCATTGGCTGTGTAA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | |
|---|
| Protein Residues | 469 |
|---|
| Protein Molecular Weight | 52218.0 |
|---|
| Protein Theoretical pI | 7.47 |
|---|
| Signalling Regions | |
|---|
| Transmembrane Regions | |
|---|
| Protein Sequence | >Serine hydroxymethyltransferase, cytosolic
MPYTLSDAHHKLITSHLVDTDPEVDSIIKDEIERQKHSIDLIASENFTSTSVFDALGTPL
SNKYSEGYPGARYYGGNEHIDRMEILCQQRALKAFHVTPDKWGVNVQTLSGSPANLQVYQ
AIMKPHERLMGLYLPDGGHLSHGYATENRKISAVSTYFESFPYRVNPETGIIDYDTLEKN
AILYRPKVLVAGTSAYCRLIDYKRMREIADKCGAYLMVDMAHISGLIAAGVIPSPFEYAD
IVTTTTHKSLRGPRGAMIFFRRGVRSINPKTGKEVLYDLENPINFSVFPGHQGGPHNHTI
AALATALKQAATPEFKEYQTQVLKNAKALESEFKNLGYRLVSNGTDSHMVLVSLREKGVD
GARVEYICEKINIALNKNSIPGDKSALVPGGVRIGAPAMTTRGMGEEDFHRIVQYINKAV
EFAQQVQQSLPKDACRLKDFKAKVDEGSDVLNTWKKEIYDWAGEYPLAV |
|---|
| References |
|---|
| External Links | |
|---|
| General Reference | - McNeil, J. B., McIntosh, E. M., Taylor, B. V., Zhang, F. R., Tang, S., Bognar, A. L. (1994). "Cloning and molecular characterization of three genes, including two genes encoding serine hydroxymethyltransferases, whose inactivation is required to render yeast auxotrophic for glycine." J Biol Chem 269:9155-9165.8132653
- Johnston, M., Hillier, L., Riles, L., Albermann, K., Andre, B., Ansorge, W., Benes, V., Bruckner, M., Delius, H., Dubois, E., Dusterhoft, A., Entian, K. D., Floeth, M., Goffeau, A., Hebling, U., Heumann, K., Heuss-Neitzel, D., Hilbert, H., Hilger, F., Kleine, K., Kotter, P., Louis, E. J., Messenguy, F., Mewes, H. W., Hoheisel, J. D., et, a. l. .. (1997). "The nucleotide sequence of Saccharomyces cerevisiae chromosome XII." Nature 387:87-90.9169871
- Norbeck, J., Blomberg, A. (1997). "Two-dimensional electrophoretic separation of yeast proteins using a non-linear wide range (pH 3-10) immobilized pH gradient in the first dimension; reproducibility and evidence for isoelectric focusing of alkaline (pI > 7) proteins." Yeast 13:1519-1534.9509572
- Ghaemmaghami, S., Huh, W. K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., O'Shea, E. K., Weissman, J. S. (2003). "Global analysis of protein expression in yeast." Nature 425:737-741.14562106
- Albuquerque, C. P., Smolka, M. B., Payne, S. H., Bafna, V., Eng, J., Zhou, H. (2008). "A multidimensional chromatography technology for in-depth phosphoproteome analysis." Mol Cell Proteomics 7:1389-1396.18407956
|
|---|



