| Identification |
|---|
| Name | Acyl-CoA desaturase 1 |
|---|
| Synonyms | - Fatty acid desaturase 1
- Stearoyl-CoA desaturase 1
|
|---|
| Gene Name | OLE1 |
|---|
| Enzyme Class | |
|---|
| Biological Properties |
|---|
| General Function | Involved in stearoyl-CoA 9-desaturase activity |
|---|
| Specific Function | Utilizes O(2) and electrons from the reduced cytochrome b(5) domain to catalyze the insertion of a double bond into a spectrum of fatty acyl-CoA substrates (Probable) |
|---|
| Cellular Location | Endoplasmic reticulum membrane; Multi-pass membrane protein (Probable) |
|---|
| SMPDB Pathways | | Biosynthesis of unsaturated fatty acids | PW002403 |  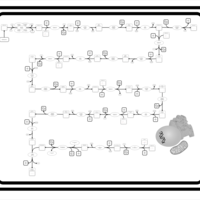 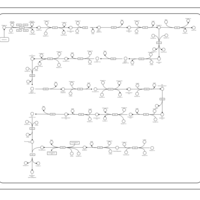 |
|
|---|
| KEGG Pathways | | Biosynthesis of unsaturated fatty acids | ec01040 |  |
|
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | |
|---|
| Metabolites | | YMDB ID | Name | View |
|---|
| YMDB00426 | NADPH | Show | | YMDB00427 | NADP | Show | | YMDB00469 | hexadec-2-enoyl-CoA | Show | | YMDB00527 | palmitoyl-CoA | Show | | YMDB00538 | stearoyl-CoA | Show | | YMDB00539 | oleoyl-CoA | Show | | YMDB00862 | hydron | Show | | YMDB00890 | water | Show | | YMDB00900 | oxygen | Show |
|
|---|
| GO Classification | | Component |
|---|
| cell part | | organelle | | membrane | | membrane-bounded organelle | | intracellular membrane-bounded organelle | | endoplasmic reticulum | | Function |
|---|
| iron ion binding | | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water | | oxidoreductase activity | | stearoyl-CoA 9-desaturase activity | | heme binding | | catalytic activity | | binding | | ion binding | | cation binding | | metal ion binding | | transition metal ion binding | | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | | Process |
|---|
| oxidation reduction | | organic acid metabolic process | | oxoacid metabolic process | | metabolic process | | carboxylic acid metabolic process | | cellular metabolic process | | lipid metabolic process | | primary metabolic process | | monocarboxylic acid metabolic process | | fatty acid metabolic process | | fatty acid biosynthetic process |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | chromosome 7 |
|---|
| Locus | YGL055W |
|---|
| Gene Sequence | >1533 bp
ATGCCAACTTCTGGAACTACTATTGAATTGATTGACGACCAATTTCCAAAGGATGACTCT
GCCAGCAGTGGCATTGTCGACGAAGTCGACTTAACGGAAGCTAATATTTTGGCTACTGGT
TTGAATAAGAAAGCACCAAGAATTGTCAACGGTTTTGGTTCTTTAATGGGCTCCAAGGAA
ATGGTTTCCGTGGAATTCGACAAGAAGGGAAACGAAAAGAAGTCCAATTTGGATCGTCTG
CTAGAAAAGGACAACCAAGAAAAAGAAGAAGCTAAAACTAAAATTCACATCTCCGAACAA
CCATGGACTTTGAATAACTGGCACCAACATTTGAACTGGTTGAACATGGTTCTTGTTTGT
GGTATGCCAATGATTGGTTGGTACTTCGCTCTCTCTGGTAAAGTACCTTTGCATTTAAAC
GTTTTCCTTTTCTCCGTTTTCTACTACGCTGTCGGTGGTGTTTCTATTACTGCCGGTTAC
CATAGATTATGGTCTCACAGATCTTACTCCGCTCACTGGCCATTGAGATTATTCTACGCT
ATCTTCGGTTGTGCTTCCGTTGAAGGGTCCGCTAAATGGTGGGGCCACTCTCACAGAATT
CACCATCGTTACACTGATACCTTGAGAGATCCTTATGACGCTCGTAGAGGTCTATGGTAC
TCCCACATGGGATGGATGCTTTTGAAGCCAAATCCAAAATACAAGGCTAGAGCTGATATT
ACCGATATGACTGATGATTGGACCATTAGATTCCAACACAGACACTACATCTTGTTGATG
TTATTAACCGCTTTCGTCATTCCAACTCTTATCTGTGGTTACTTTTTCAACGACTATATG
GGTGGTTTGATCTATGCCGGTTTTATTCGTGTCTTTGTCATTCAACAAGCTACCTTTTGC
ATTAACTCCATGGCTCATTACATCGGTACCCAACCATTCGATGACAGAAGAACCCCTCGT
GACAACTGGATTACTGCCATTGTTACTTTCGGTGAAGGTTACCATAACTTCCACCACGAA
TTCCCAACTGATTACAGAAACGCTATTAAGTGGTACCAATACGACCCAACTAAGGTTATC
ATCTATTTGACTTCTTTAGTTGGTCTAGCATACGACTTGAAGAAATTCTCTCAAAATGCT
ATTGAAGAAGCCTTGATTCAACAAGAACAAAAGAAGATCAATAAAAAGAAGGCTAAGATT
AACTGGGGTCCAGTTTTGACTGATTTGCCAATGTGGGACAAACAAACCTTCTTGGCTAAG
TCTAAGGAAAACAAGGGTTTGGTTATCATTTCTGGTATTGTTCACGACGTATCTGGTTAT
ATCTCTGAACATCCAGGTGGTGAAACTTTAATTAAAACTGCATTAGGTAAGGACGCTACC
AAGGCTTTCAGTGGTGGTGTCTACCGTCACTCAAATGCCGCTCAAAATGTCTTGGCTGAT
ATGAGAGTGGCTGTTATCAAGGAAAGTAAGAACTCTGCTATTAGAATGGCTAGTAAGAGA
GGTGAAATCTACGAAACTGGTAAGTTCTTTTAA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | |
|---|
| Protein Residues | 510 |
|---|
| Protein Molecular Weight | 58402.60156 |
|---|
| Protein Theoretical pI | 9.33 |
|---|
| Signalling Regions | |
|---|
| Transmembrane Regions | - 113-133
- 139-159
- 256-276
- 281-301
|
|---|
| Protein Sequence | >Acyl-CoA desaturase 1
MPTSGTTIELIDDQFPKDDSASSGIVDEVDLTEANILATGLNKKAPRIVNGFGSLMGSKE
MVSVEFDKKGNEKKSNLDRLLEKDNQEKEEAKTKIHISEQPWTLNNWHQHLNWLNMVLVC
GMPMIGWYFALSGKVPLHLNVFLFSVFYYAVGGVSITAGYHRLWSHRSYSAHWPLRLFYA
IFGCASVEGSAKWWGHSHRIHHRYTDTLRDPYDARRGLWYSHMGWMLLKPNPKYKARADI
TDMTDDWTIRFQHRHYILLMLLTAFVIPTLICGYFFNDYMGGLIYAGFIRVFVIQQATFC
INSLAHYIGTQPFDDRRTPRDNWITAIVTFGEGYHNFHHEFPTDYRNAIKWYQYDPTKVI
IYLTSLVGLAYDLKKFSQNAIEEALIQQEQKKINKKKAKINWGPVLTDLPMWDKQTFLAK
SKENKGLVIISGIVHDVSGYISEHPGGETLIKTALGKDATKAFSGGVYRHSNAAQNVLAD
MRVAVIKESKNSAIRMASKRGEIYETGKFF |
|---|
| References |
|---|
| External Links | |
|---|
| General Reference | - Stukey, J. E., McDonough, V. M., Martin, C. E. (1990). "The OLE1 gene of Saccharomyces cerevisiae encodes the delta 9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene." J Biol Chem 265:20144-20149.1978720
- Feuermann, M., de Montigny, J., Potier, S., Souciet, J. L. (1997). "The characterization of two new clusters of duplicated genes suggests a 'Lego' organization of the yeast Saccharomyces cerevisiae chromosomes." Yeast 13:861-869.9234674
- Tettelin, H., Agostoni Carbone, M. L., Albermann, K., Albers, M., Arroyo, J., Backes, U., Barreiros, T., Bertani, I., Bjourson, A. J., Bruckner, M., Bruschi, C. V., Carignani, G., Castagnoli, L., Cerdan, E., Clemente, M. L., Coblenz, A., Coglievina, M., Coissac, E., Defoor, E., Del Bino, S., Delius, H., Delneri, D., de Wergifosse, P., Dujon, B., Kleine, K., et, a. l. .. (1997). "The nucleotide sequence of Saccharomyces cerevisiae chromosome VII." Nature 387:81-84.9169869
- Kim, H., Melen, K., Osterberg, M., von Heijne, G. (2006). "A global topology map of the Saccharomyces cerevisiae membrane proteome." Proc Natl Acad Sci U S A 103:11142-11147.16847258
|
|---|
