| Identification |
|---|
| Name | Glucokinase-1 |
|---|
| Synonyms | |
|---|
| Gene Name | GLK1 |
|---|
| Enzyme Class | |
|---|
| Biological Properties |
|---|
| General Function | Involved in ATP binding |
|---|
| Specific Function | Two isoenzymes, hexokinase-1 and hexokinase-2, can phosphorylate keto- and aldohexoses in yeast, whereas a third isoenzyme, GLK, is specific for aldohexoses. All glucose phosphorylating enzymes are involved in glucose uptake |
|---|
| Cellular Location | Not Available |
|---|
| SMPDB Pathways | | Amino sugar and nucleotide sugar metabolism | PW002413 | 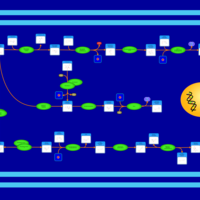 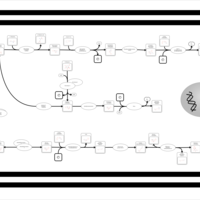 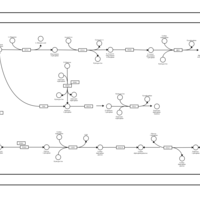 | | Starch and sucrose metabolism | PW002481 | 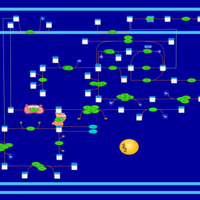 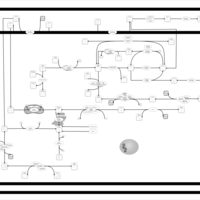 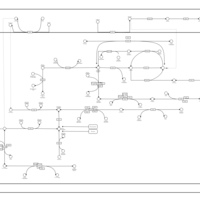 |
|
|---|
| KEGG Pathways | | Amino sugar and nucleotide sugar metabolism | ec00520 |  | | Galactose metabolism | ec00052 |  | | Glycolysis / Gluconeogenesis | ec00010 |  | | Starch and sucrose metabolism | ec00500 |  |
|
|---|
| SMPDB Reactions | |
|---|
| KEGG Reactions | |
|---|
| Metabolites | | YMDB ID | Name | View |
|---|
| YMDB00109 | Adenosine triphosphate | Show | | YMDB00242 | Mannose 6-phosphate | Show | | YMDB00286 | D-Glucose | Show | | YMDB00311 | Beta-D-Glucose 6-phosphate | Show | | YMDB00582 | aldehydo-D-glucose 6-phosphate | Show | | YMDB00657 | D-Fructose | Show | | YMDB00862 | hydron | Show | | YMDB00894 | D-Mannose | Show | | YMDB00914 | ADP | Show | | YMDB00947 | Glucose 6-phosphate | Show | | YMDB16176 | Beta-D-Fructose 6-phosphate | Show |
|
|---|
| GO Classification | | Component |
|---|
| Not Available | | Function |
|---|
| catalytic activity | | transferase activity | | hexokinase activity | | transferase activity, transferring phosphorus-containing groups | | binding | | nucleoside binding | | purine nucleoside binding | | adenyl nucleotide binding | | adenyl ribonucleotide binding | | ATP binding | | phosphotransferase activity, alcohol group as acceptor | | Process |
|---|
| monosaccharide metabolic process | | hexose metabolic process | | metabolic process | | glucose metabolic process | | glucose catabolic process | | glycolysis | | primary metabolic process | | carbohydrate metabolic process | | small molecule metabolic process | | alcohol metabolic process |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | chromosome 3 |
|---|
| Locus | YCL040W |
|---|
| Gene Sequence | >1503 bp
ATGTCATTCGACGACTTACACAAAGCCACTGAGAGAGCGGTCATCCAGGCCGTGGACCAG
ATCTGCGACGATTTCGAGGTTACCCCCGAGAAGCTGGACGAATTAACTGCTTACTTCATC
GAACAAATGGAAAAAGGTCTAGCTCCACCAAAGGAAGGCCACACATTGGCCTCGGACAAA
GGTCTTCCTATGATTCCGGCGTTCGTCACCGGGTCACCCAACGGGACGGAGCGCGGTGTT
TTACTAGCCGCCGACCTGGGTGGTACCAATTTCCGTATATGTTCTGTTAACTTGCATGGA
GATCATACTTTCTCCATGGAGCAAATGAAGTCCAAGATTCCCGATGATTTGCTAGACGAT
GAGAACGTCACATCTGACGACCTGTTTGGGTTTCTAGCACGTCGTACACTGGCCTTTATG
AAGAAGTATCACCCGGACGAGTTGGCCAAGGGTAAAGACGCCAAGCCCATGAAACTGGGG
TTCACTTTCTCATACCCTGTAGACCAGACCTCTCTAAACTCCGGGACATTGATCCGTTGG
ACCAAGGGTTTCCGCATCGCGGACACCGTCGGAAAGGATGTCGTGCAATTGTACCAGGAG
CAATTAAGCGCTCAGGGTATGCCTATGATCAAGGTTGTTGCATTAACCAACGACACCGTC
GGAACGTACCTATCGCATTGCTACACGTCCGATAACACGGACTCAATGACGTCCGGAGAA
ATCTCGGAGCCGGTCATCGGATGTATTTTCGGTACCGGTACCAATGGGTGCTATATGGAG
GAGATCAACAAGATCACGAAGTTGCCACAGGAGTTGCGTGACAAGTTGATAAAGGAGGGT
AAGACACACATGATCATCAATGTCGAATGGGGGTCCTTCGATAATGAGCTCAAGCACTTG
CCTACTACTAAGTATGACGTCGTAATTGACCAGAAACTGTCAACGAACCCGGGATTTCAC
TTGTTTGAAAAACGTGTCTCAGGGATGTTCTTGGGTGAGGTGTTGCGTAACATTTTAGTG
GACTTGCACTCGCAAGGCTTGCTTTTGCAACAGTACAGGTCCAAGGAACAACTTCCTCGC
CACTTGACTACACCTTTCCAGTTGTCATCCGAAGTGCTGTCGCATATTGAAATTGACGAC
TCGACAGGTCTACGTGAAACAGAGTTGTCATTATTACAGAGTCTCAGACTGCCCACCACT
CCAACAGAGCGTGTTCAAATTCAAAAATTGGTGCGCGCGATTTCTAGGAGATCTGCGTAT
TTAGCCGCCGTGCCGCTTGCCGCGATATTGATCAAGACAAATGCTTTGAACAAGAGATAT
CATGGTGAAGTCGAGATCGGTTGTGATGGTTCCGTTGTGGAATACTACCCCGGTTTCAGA
TCTATGCTGAGACACGCCTTAGCCTTGTCACCCTTGGGTGCCGAGGGTGAGAGGAAGGTG
CACTTGAAGATTGCCAAGGATGGTTCCGGAGTGGGTGCCGCCTTGTGTGCGCTTGTAGCA
TGA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | |
|---|
| Protein Residues | 500 |
|---|
| Protein Molecular Weight | 55376.89844 |
|---|
| Protein Theoretical pI | 6.13 |
|---|
| Signalling Regions | |
|---|
| Transmembrane Regions | |
|---|
| Protein Sequence | >Glucokinase-1
MSFDDLHKATERAVIQAVDQICDDFEVTPEKLDELTAYFIEQMEKGLAPPKEGHTLASDK
GLPMIPAFVTGSPNGTERGVLLAADLGGTNFRICSVNLHGDHTFSMEQMKSKIPDDLLDD
ENVTSDDLFGFLARRTLAFMKKYHPDELAKGKDAKPMKLGFTFSYPVDQTSLNSGTLIRW
TKGFRIADTVGKDVVQLYQEQLSAQGMPMIKVVALTNDTVGTYLSHCYTSDNTDSMTSGE
ISEPVIGCIFGTGTNGCYMEEINKITKLPQELRDKLIKEGKTHMIINVEWGSFDNELKHL
PTTKYDVVIDQKLSTNPGFHLFEKRVSGMFLGEVLRNILVDLHSQGLLLQQYRSKEQLPR
HLTTPFQLSSEVLSHIEIDDSTGLRETELSLLQSLRLPTTPTERVQIQKLVRAISRRSAY
LAAVPLAAILIKTNALNKRYHGEVEIGCDGSVVEYYPGFRSMLRHALALSPLGAEGERKV
HLKIAKDGSGVGAALCALVA |
|---|
| References |
|---|
| External Links | |
|---|
| General Reference | - Albig, W., Entian, K. D. (1988). "Structure of yeast glucokinase, a strongly diverged specific aldo-hexose-phosphorylating isoenzyme." Gene 73:141-152.3072253
- Scherens, B., Messenguy, F., Gigot, D., Dubois, E. (1992). "The complete sequence of a 9,543 bp segment on the left arm of chromosome III reveals five open reading frames including glucokinase and the protein disulfide isomerase." Yeast 8:577-585.1523890
- Oliver, S. G., van der Aart, Q. J., Agostoni-Carbone, M. L., Aigle, M., Alberghina, L., Alexandraki, D., Antoine, G., Anwar, R., Ballesta, J. P., Benit, P., et, a. l. .. (1992). "The complete DNA sequence of yeast chromosome III." Nature 357:38-46.1574125
- Ghaemmaghami, S., Huh, W. K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., O'Shea, E. K., Weissman, J. S. (2003). "Global analysis of protein expression in yeast." Nature 425:737-741.14562106
- Albuquerque, C. P., Smolka, M. B., Payne, S. H., Bafna, V., Eng, J., Zhou, H. (2008). "A multidimensional chromatography technology for in-depth phosphoproteome analysis." Mol Cell Proteomics 7:1389-1396.18407956
|
|---|



