| Identification |
|---|
| Name | Thiamine biosynthetic bifunctional enzyme |
|---|
| Synonyms | - Thiamine-phosphate pyrophosphorylase
- TMP pyrophosphorylase
- TMP-PPASE
- Hydroxyethylthiazole kinase
- 4-methyl-5-beta-hydroxyethylthiazole kinase
- TH kinase
- THZ kinase
|
|---|
| Gene Name | THI6 |
|---|
| Enzyme Class | |
|---|
| Biological Properties |
|---|
| General Function | Involved in catalytic activity |
|---|
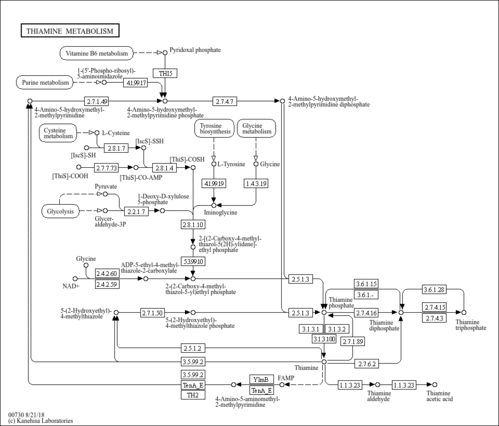
| Specific Function | Essential for thiamine biosynthesis. The kinase activity is involved in the salvage synthesis of TH-P from the thiazole |
|---|
| Cellular Location | Not Available |
|---|
| SMPDB Pathways | |
|---|
| KEGG Pathways | |
|---|
| SMPDB Reactions | |
|---|
| KEGG Reactions | |
|---|
| Metabolites | | YMDB ID | Name | View |
|---|
| YMDB00109 | Adenosine triphosphate | Show | | YMDB00117 | 4-amino-2-methyl-5-diphosphomethylpyrimidine | Show | | YMDB00164 | Thiamine monophosphate | Show | | YMDB00219 | Pyrophosphate | Show | | YMDB00319 | 4-Methyl-5-(2-phosphonooxyethyl)thiazole | Show | | YMDB00706 | 5-(2-Hydroxyethyl)-4-methylthiazole | Show | | YMDB00774 | Thiamine(1+) monophosphate | Show | | YMDB00862 | hydron | Show | | YMDB00914 | ADP | Show |
|
|---|
| GO Classification | | Component |
|---|
| Not Available | | Function |
|---|
| transferase activity, transferring alkyl or aryl (other than methyl) groups | | thiamin-phosphate diphosphorylase activity | | hydroxyethylthiazole kinase activity | | catalytic activity | | transferase activity | | transferase activity, transferring phosphorus-containing groups | | kinase activity | | Process |
|---|
| thiamin and derivative metabolic process | | thiamin metabolic process | | thiamin biosynthetic process | | metabolic process | | cellular metabolic process | | cellular aromatic compound metabolic process |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | chromosome 16 |
|---|
| Locus | YPL214C |
|---|
| Gene Sequence | >1623 bp
ATGGTATTTACTAAGGAAGAAGTTGATTACTCATTATATCTGGTTACAGATTCTACCATG
CTTCCACCGGGAACTACTTTATGTTCTCAGGTTGAAGCTGGGTTGAAAAATGGTGTAACG
CTAGTTCAAATCCGTGAAAAGGATATTGAAACAAAAAATTTCGTTGCAGAGGCTTTAGAA
GTTCAAAAAATATGTAAGAAGTACAATGTTCCACTTATTATCAATGACCGTATAGACGTC
GCTATGGCAATTGACGCCGATGGGGTTCATGTGGGCCAGGACGATATGCCAATCCCAATG
GTAAGAAAACTTTTGGGCCCTTCTAAAATTCTTGGATGGAGTGTTGGTAAACCTTCGGAG
GTAGAGACATTGGCTAAATGGGGGCCAGATATGGTAGATTATATTGGTGTTGGTACTCTT
TTCCCTACATCAACAAAGAAAAATCCTAAGAAATCACCCATGGGTCCTCAAGGCGCCATT
GCCATTTTGGATGCTTTGGAGGAATTTAAAGCTACATGGTGTAGAACCGTTGGTATTGGT
GGTCTTCATCCTGACAATATTCAACGTGTGCTTTGTCAATGTGTTGCTTCAAATGGAAAG
AGATCGTTGGATGGTATCTCTCTAGTCAGCGATATTATGGCCGCTCCAGATGCATGTGCA
GCAACCAAGAGGTTGAGAGGCTTGTTAGATGCTACCAGATACCAATTTGTTGAGTGCGAG
TTAAATAATACATTTCCGACCACAACTTCAATTCAAAACGTTATATCTCAAGTATCAAAC
AATCGTCCATTAGTGCAACATATCACCAATAAGGTTCATCAAAATTTTGGTGCCAATGTC
ACTCTAGCTTTAGGCTCATCTCCTATCATGTCTGAAATTGAAAGTGAAGTATCTGAACTA
GCAAGAATCCCAAATGCTTCTTTACTGTTAAATACCGGATCAGTGGCACCTATTGAAATG
CTAAAAGCCGCAATTAATGCTTATAATGAAGTAAATAGACCCATCACCTTTGACCCGGTC
GGGTACAGCGCCACTGAAACAAGGCTCTGTCTAAACAACACTTTGCTCACTTACGGCCAA
TTTGCTTGTATAAAGGGCAATTGCAGTGAAATACTGTCTCTAGCCAAGTTAAATAACCAT
AAAATGAAGGGCGTCGACTCTAGCAGCGGCAAAACGAACATCGACACACTTGTGCGCGCT
ACACAAATTGTGGCATTCCAATACAGGACTGTTGCTGTTTGCACGGGTGAGTTTGATTGC
GTTGCTGATGGTACTTTTGGAGGCGAATACAAACTTTCATCAGGAACAGAAGGAATAACC
GCTGAAGATCTTCCATGTGTAATAATTGAAGATGGCCCAATTCCTATCATGGGTGACATC
ACTGCAAGTGGATGTTCACTTGGTTCTACAATTGCTTCCTTTATCGGTGGGTTAGATTCC
ACTGGAAAACTGTTTGATGCTGTGGTTGGTGCTGTCCTATTATACAAATCAGCTGGTAAG
TTGGCTTCCACCCGTTGTCAAGGAAGTGGTTCATTCCATGTCGAATTGATTGACGCATTA
TATCAATTATTCCATGAAAATAAACCAGAAAAGTGGTCCGCTTCTTTGAAGAAATTCAAA
TAA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | |
|---|
| Protein Residues | 540 |
|---|
| Protein Molecular Weight | 58058.19922 |
|---|
| Protein Theoretical pI | 5.74 |
|---|
| Signalling Regions | |
|---|
| Transmembrane Regions | |
|---|
| Protein Sequence | >Thiamine biosynthetic bifunctional enzyme
MVFTKEEVDYSLYLVTDSTMLPPGTTLCSQVEAGLKNGVTLVQIREKDIETKNFVAEALE
VQKICKKYNVPLIINDRIDVAMAIDADGVHVGQDDMPIPMVRKLLGPSKILGWSVGKPSE
VETLAKWGPDMVDYIGVGTLFPTSTKKNPKKSPMGPQGAIAILDALEEFKATWCRTVGIG
GLHPDNIQRVLCQCVASNGKRSLDGISLVSDIMAAPDACAATKRLRGLLDATRYQFVECE
LNNTFPTTTSIQNVISQVSNNRPLVQHITNKVHQNFGANVTLALGSSPIMSEIESEVSEL
ARIPNASLLLNTGSVAPIEMLKAAINAYNEVNRPITFDPVGYSATETRLCLNNTLLTYGQ
FACIKGNCSEILSLAKLNNHKMKGVDSSSGKTNIDTLVRATQIVAFQYRTVAVCTGEFDC
VADGTFGGEYKLSSGTEGITAEDLPCVIIEDGPIPIMGDITASGCSLGSTIASFIGGLDS
TGKLFDAVVGAVLLYKSAGKLASTRCQGSGSFHVELIDALYQLFHENKPEKWSASLKKFK
|
|---|
| References |
|---|
| External Links | |
|---|
| General Reference | - Nosaka, K., Nishimura, H., Kawasaki, Y., Tsujihara, T., Iwashima, A. (1994). "Isolation and characterization of the THI6 gene encoding a bifunctional thiamin-phosphate pyrophosphorylase/hydroxyethylthiazole kinase from Saccharomyces cerevisiae." J Biol Chem 269:30510-30516.7982968
- Bussey, H., Storms, R. K., Ahmed, A., Albermann, K., Allen, E., Ansorge, W., Araujo, R., Aparicio, A., Barrell, B., Badcock, K., Benes, V., Botstein, D., Bowman, S., Bruckner, M., Carpenter, J., Cherry, J. M., Chung, E., Churcher, C., Coster, F., Davis, K., Davis, R. W., Dietrich, F. S., Delius, H., DiPaolo, T., Hani, J., et, a. l. .. (1997). "The nucleotide sequence of Saccharomyces cerevisiae chromosome XVI." Nature 387:103-105.9169875
|
|---|