L-aminoadipate-semialdehyde dehydrogenase (P07702)
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | L-aminoadipate-semialdehyde dehydrogenase | |||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| |||||||||||||||||||||||||||||||||||||||||||||
| Gene Name | LYS2 | |||||||||||||||||||||||||||||||||||||||||||||
| Enzyme Class | ||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||
| General Function | Involved in ligase activity | |||||||||||||||||||||||||||||||||||||||||||||
| Specific Function | Catalyzes the activation of alpha-aminoadipate by ATP- dependent adenylation and the reduction of activated alpha- aminoadipate by NADPH | |||||||||||||||||||||||||||||||||||||||||||||
| Cellular Location | Not Available | |||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways |
| |||||||||||||||||||||||||||||||||||||||||||||
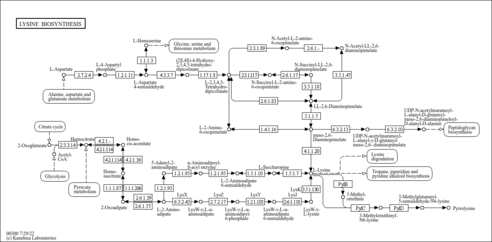
| KEGG Pathways |
| |||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Reactions |
| |||||||||||||||||||||||||||||||||||||||||||||
| KEGG Reactions | Not Available | |||||||||||||||||||||||||||||||||||||||||||||
| Metabolites |
| |||||||||||||||||||||||||||||||||||||||||||||
| GO Classification |
| |||||||||||||||||||||||||||||||||||||||||||||
| Gene Properties | ||||||||||||||||||||||||||||||||||||||||||||||
| Chromosome Location | chromosome 2 | |||||||||||||||||||||||||||||||||||||||||||||
| Locus | YBR115C | |||||||||||||||||||||||||||||||||||||||||||||
| Gene Sequence | >4179 bp ATGACTAACGAAAAGGTCTGGATAGAGAAGTTGGATAATCCAACTCTTTCAGTGTTACCA CATGACTTTTTACGCCCACAACAAGAACCTTATACGAAACAAGCTACATATTCGTTACAG CTACCTCAGCTCGATGTGCCTCATGATAGTTTTTCTAACAAATACGCTGTCGCTTTGAGT GTATGGGCTGCATTGATATATAGAGTAACCGGTGACGATGATATTGTTCTTTATATTGCG AATAACAAAATCTTAAGATTCAATATTCAACCAACGTGGTCATTTAATGAGCTGTATTCT ACAATTAACAATGAGTTGAACAAGCTCAATTCTATTGAGGCCAATTTTTCCTTTGACGAG CTAGCTGAAAAAATTCAAAGTTGCCAAGATCTGGAAAGGACCCCTCAGTTGTTCCGTTTG GCCTTTTTGGAAAACCAAGATTTCAAATTAGACGAGTTCAAGCATCATTTAGTGGACTTT GCTTTGAATTTGGATACCAGTAATAATGCGCATGTTTTGAACTTAATTTATAACAGCTTA CTGTATTCGAATGAAAGAGTAACCATTGTTGCGGACCAATTTACTCAATATTTGACTGCT GCGCTAAGCGATCCATCCAATTGCATAACTAAAATCTCTCTGATCACCGCATCATCCAAG GATAGTTTACCTGATCCAACTAAGAACTTGGGCTGGTGCGATTTCGTGGGGTGTATTCAC GACATTTTCCAGGACAATGCTGAAGCCTTCCCAGAGAGAACCTGTGTTGTGGAGACTCCA ACACTAAATTCCGACAAGTCCCGTTCTTTCACTTATCGCGACATCAACCGCACTTCTAAC ATAGTTGCCCATTATTTGATTAAAACAGGTATCAAAAGAGGTGATGTAGTGATGATCTAT TCTTCTAGGGGTGTGGATTTGATGGTATGTGTGATGGGTGTCTTGAAAGCCGGCGCAACC TTTTCAGTTATCGACCCTGCATATCCCCCAGCCAGACAAACCATTTACTTAGGTGTTGCT AAACCACGTGGGTTGATTGTTATTAGAGCTGCTGGACAATTGGATCAACTAGTAGAAGAT TACATCAATGATGAATTGGAGATTGTTTCAAGAATCAATTCCATCGCTATTCAAGAAAAT GGTACCATTGAAGGTGGCAAATTGGACAATGGCGAGGATGTTTTGGCTCCATATGATCAC TACAAAGACACCAGAACAGGTGTTGTAGTTGGACCAGATTCCAACCCAACCCTATCTTTC ACATCTGGTTCCGAAGGTATTCCTAAGGGTGTTCTTGGTAGACATTTTTCCTTGGCTTAT TATTTCAATTGGATGTCCAAAAGGTTCAACTTAACAGAAAATGATAAATTCACAATGCTG AGCGGTATTGCACATGATCCAATTCAAAGAGATATGTTTACACCATTATTTTTAGGTGCC CAATTGTATGTCCCTACTCAAGATGATATTGGTACACCGGGCCGTTTAGCGGAATGGATG AGTAAGTATGGTTGCACAGTTACCCATTTAACACCTGCCATGGGTCAATTACTTACTGCC CAAGCTACTACACCATTCCCTAAGTTACATCATGCGTTCTTTGTGGGTGACATTTTAACA AAACGTGATTGTCTGAGGTTACAAACCTTGGCAGAAAATTGCCGTATTGTTAATATGTAC GGTACCACTGAAACACAGCGTGCAGTTTCTTATTTCGAAGTTAAATCAAAAAATGACGAT CCAAACTTTTTGAAAAAATTGAAAGATGTCATGCCTGCTGGTAAAGGTATGTTGAACGTT CAGCTACTAGTTGTTAACAGGAACGATCGTACTCAAATATGTGGTATTGGCGAAATAGGT GAGATTTATGTTCGTGCAGGTGGTTTGGCCGAAGGTTATAGAGGATTACCAGAATTGAAT AAAGAAAAATTTGTGAACAACTGGTTTGTTGAAAAAGATCACTGGAATTATTTGGATAAG GATAATGGTGAACCTTGGAGACAATTCTGGTTAGGTCCAAGAGATAGATTGTACAGAACG GGTGATTTAGGTCGTTATCTACCAAACGGTGACTGTGAATGTTGCGGTAGGGCTGATGAT CAAGTTAAAATTCGTGGGTTCAGAATCGAATTAGGAGAAATAGATACGCACATTTCCCAA CATCCATTGGTAAGAGAAAACATTACTTTAGTTCGCAAAAATGCCGACAATGAGCCAACA TTGATCACATTTATGGTCCCAAGATTTGACAAGCCAGATGACTTGTCTAAGTTCCAAAGT GATGTTCCAAAGGAGGTTGAAACTGACCCTATAGTTAAGGGCTTAATCGGTTACCATCTT TTATCCAAGGACATCAGGACTTTCTTAAAGAAAAGATTGGCTAGCTATGCTATGCCTTCC TTGATTGTGGTTATGGATAAACTACCATTGAATCCAAATGGTAAAGTTGATAAGCCTAAA CTTCAATTCCCAACTCCCAAGCAATTAAATTTGGTAGCTGAAAATACAGTTTCTGAAACT GACGACTCTCAGTTTACCAATGTTGAGCGCGAGGTTAGAGACTTATGGTTAAGTATATTA CCTACCAAGCCAGCATCTGTATCACCAGATGATTCGTTTTTCGATTTAGGTGGTCATTCT ATCTTGGCTACCAAAATGATTTTTACCTTAAAGAAAAAGCTGCAAGTTGATTTACCATTG GGCACAATTTTCAAGTATCCAACGATAAAGGCCTTTGCCGCGGAAATTGACAGAATTAAA TCATCGGGTGGATCATCTCAAGGTGAGGTCGTCGAAAATGTCACTGCAAATTATGCGGAA GACGCCAAGAAATTGGTTGAGACGCTACCAAGTTCGTACCCCTCTCGAGAATATTTTGTT GAACCTAATAGTGCCGAAGGAAAAACAACAATTAATGTGTTTGTTACCGGTGTCACAGGA TTTCTGGGCTCCTACATCCTTGCAGATTTGTTAGGACGTTCTCCAAAGAACTACAGTTTC AAAGTGTTTGCCCACGTCAGGGCCAAGGATGAAGAAGCTGCATTTGCAAGATTACAAAAG GCAGGTATCACCTATGGTACTTGGAACGAAAAATTTGCCTCAAATATTAAAGTTGTATTA GGCGATTTATCTAAAAGCCAATTTGGTCTTTCAGATGAGAAGTGGATGGATTTGGCAAAC ACAGTTGATATAATTATCCATAATGGTGCGTTAGTTCACTGGGTTTATCCATATGCCAAA TTGAGGGATCCAAATGTTATTTCAACTATCAATGTTATGAGCTTAGCCGCCGTCGGCAAG CCAAAGTTCTTTGACTTTGTTTCCTCCACTTCTACTCTTGACACTGAATACTACTTTAAT TTGTCAGATAAACTTGTTAGCGAAGGGAAGCCAGGCATTTTAGAATCAGACGATTTAATG AACTCTGCAAGCGGGCTCACTGGTGGATATGGTCAGTCCAAATGGGCTGCTGAGTACATC ATTAGACGTGCAGGTGAAAGGGGCCTACGTGGGTGTATTGTCAGACCAGGTTACGTAACA GGTGCCTCTGCCAATGGTTCTTCAAACACAGATGATTTCTTATTGAGATTTTTGAAAGGT TCAGTCCAATTAGGTAAGATTCCAGATATCGAAAATTCCGTGAATATGGTTCCAGTAGAT CATGTTGCTCGTGTTGTTGTTGCTACGTCTTTGAATCCTCCCAAAGAAAATGAATTGGCC GTTGCTCAAGTAACGGGTCACCCAAGAATATTATTCAAAGACTACTTGTATACTTTACAC GATTATGGTTACGATGTCGAAATCGAAAGCTATTCTAAATGGAAGAAATCATTGGAGGCG TCTGTTATTGACAGGAATGAAGAAAATGCGTTGTATCCTTTGCTACACATGGTCTTAGAC AACTTACCTGAAAGTACCAAAGCTCCGGAACTAGACGATAGGAACGCCGTGGCATCTTTA AAGAAAGACACCGCATGGACAGGTGTTGATTGGTCTAATGGAATAGGTGTTACTCCAGAA GAGGTTGGTATATATATTGCATTTTTAAACAAGGTTGGATTTTTACCTCCACCAACTCAT AATGACAAACTTCCACTGCCAAGTATAGAACTAACTCAAGCGCAAATAAGTCTAGTTGCT TCAGGTGCTGGTGCTCGTGGAAGCTCCGCAGCAGCTTAA | |||||||||||||||||||||||||||||||||||||||||||||
| Protein Properties | ||||||||||||||||||||||||||||||||||||||||||||||
| Pfam Domain Function | ||||||||||||||||||||||||||||||||||||||||||||||
| Protein Residues | 1392 | |||||||||||||||||||||||||||||||||||||||||||||
| Protein Molecular Weight | 155344.0 | |||||||||||||||||||||||||||||||||||||||||||||
| Protein Theoretical pI | 5.75 | |||||||||||||||||||||||||||||||||||||||||||||
| Signalling Regions |
| |||||||||||||||||||||||||||||||||||||||||||||
| Transmembrane Regions |
| |||||||||||||||||||||||||||||||||||||||||||||
| Protein Sequence | >L-aminoadipate-semialdehyde dehydrogenase MTNEKVWIEKLDNPTLSVLPHDFLRPQQEPYTKQATYSLQLPQLDVPHDSFSNKYAVALS VWAALIYRVTGDDDIVLYIANNKILRFNIQPTWSFNELYSTINNELNKLNSIEANFSFDE LAEKIQSCQDLERTPQLFRLAFLENQDFKLDEFKHHLVDFALNLDTSNNAHVLNLIYNSL LYSNERVTIVADQFTQYLTAALSDPSNCITKISLITASSKDSLPDPTKNLGWCDFVGCIH DIFQDNAEAFPERTCVVETPTLNSDKSRSFTYRDINRTSNIVAHYLIKTGIKRGDVVMIY SSRGVDLMVCVMGVLKAGATFSVIDPAYPPARQTIYLGVAKPRGLIVIRAAGQLDQLVED YINDELEIVSRINSIAIQENGTIEGGKLDNGEDVLAPYDHYKDTRTGVVVGPDSNPTLSF TSGSEGIPKGVLGRHFSLAYYFNWMSKRFNLTENDKFTMLSGIAHDPIQRDMFTPLFLGA QLYVPTQDDIGTPGRLAEWMSKYGCTVTHLTPAMGQLLTAQATTPFPKLHHAFFVGDILT KRDCLRLQTLAENCRIVNMYGTTETQRAVSYFEVKSKNDDPNFLKKLKDVMPAGKGMLNV QLLVVNRNDRTQICGIGEIGEIYVRAGGLAEGYRGLPELNKEKFVNNWFVEKDHWNYLDK DNGEPWRQFWLGPRDRLYRTGDLGRYLPNGDCECCGRADDQVKIRGFRIELGEIDTHISQ HPLVRENITLVRKNADNEPTLITFMVPRFDKPDDLSKFQSDVPKEVETDPIVKGLIGYHL LSKDIRTFLKKRLASYAMPSLIVVMDKLPLNPNGKVDKPKLQFPTPKQLNLVAENTVSET DDSQFTNVEREVRDLWLSILPTKPASVSPDDSFFDLGGHSILATKMIFTLKKKLQVDLPL GTIFKYPTIKAFAAEIDRIKSSGGSSQGEVVENVTANYAEDAKKLVETLPSSYPSREYFV EPNSAEGKTTINVFVTGVTGFLGSYILADLLGRSPKNYSFKVFAHVRAKDEEAAFARLQK AGITYGTWNEKFASNIKVVLGDLSKSQFGLSDEKWMDLANTVDIIIHNGALVHWVYPYAK LRDPNVISTINVMSLAAVGKPKFFDFVSSTSTLDTEYYFNLSDKLVSEGKPGILESDDLM NSASGLTGGYGQSKWAAEYIIRRAGERGLRGCIVRPGYVTGASANGSSNTDDFLLRFLKG SVQLGKIPDIENSVNMVPVDHVARVVVATSLNPPKENELAVAQVTGHPRILFKDYLYTLH DYGYDVEIESYSKWKKSLEASVIDRNEENALYPLLHMVLDNLPESTKAPELDDRNAVASL KKDTAWTGVDWSNGIGVTPEEVGIYIAFLNKVGFLPPPTHNDKLPLPSIELTQAQISLVA SGAGARGSSAAA | |||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||
| External Links |
| |||||||||||||||||||||||||||||||||||||||||||||
| General Reference |
| |||||||||||||||||||||||||||||||||||||||||||||