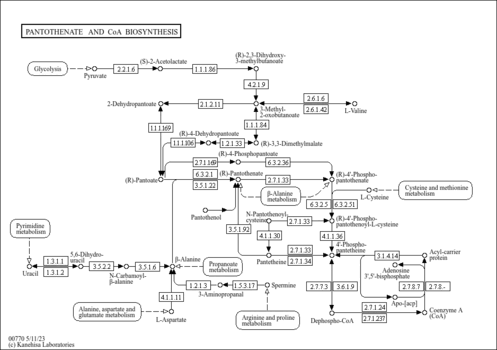
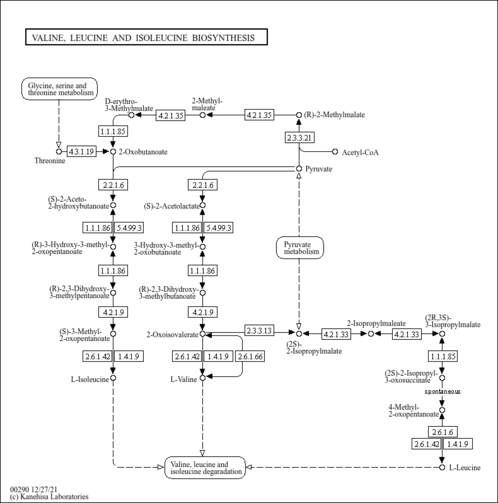
| General Reference | - Petersen, J. G., Holmberg, S. (1986). "The ILV5 gene of Saccharomyces cerevisiae is highly expressed." Nucleic Acids Res 14:9631-9651.3027658
- Johnston, M., Hillier, L., Riles, L., Albermann, K., Andre, B., Ansorge, W., Benes, V., Bruckner, M., Delius, H., Dubois, E., Dusterhoft, A., Entian, K. D., Floeth, M., Goffeau, A., Hebling, U., Heumann, K., Heuss-Neitzel, D., Hilbert, H., Hilger, F., Kleine, K., Kotter, P., Louis, E. J., Messenguy, F., Mewes, H. W., Hoheisel, J. D., et, a. l. .. (1997). "The nucleotide sequence of Saccharomyces cerevisiae chromosome XII." Nature 387:87-90.9169871
- Garrels, J. I., Futcher, B., Kobayashi, R., Latter, G. I., Schwender, B., Volpe, T., Warner, J. R., McLaughlin, C. S. (1994). "Protein identifications for a Saccharomyces cerevisiae protein database." Electrophoresis 15:1466-1486.7895733
- Shevchenko, A., Jensen, O. N., Podtelejnikov, A. V., Sagliocco, F., Wilm, M., Vorm, O., Mortensen, P., Shevchenko, A., Boucherie, H., Mann, M. (1996). "Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels." Proc Natl Acad Sci U S A 93:14440-14445.8962070
- Ghaemmaghami, S., Huh, W. K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., O'Shea, E. K., Weissman, J. S. (2003). "Global analysis of protein expression in yeast." Nature 425:737-741.14562106
- Albuquerque, C. P., Smolka, M. B., Payne, S. H., Bafna, V., Eng, J., Zhou, H. (2008). "A multidimensional chromatography technology for in-depth phosphoproteome analysis." Mol Cell Proteomics 7:1389-1396.18407956
|
|---|