| Identification |
|---|
| Name | CRE-binding bZIP protein SKO1 |
|---|
| Synonyms | Not Available |
|---|
| Gene Name | SKO1 |
|---|
| Enzyme Class | Not Available |
|---|
| Biological Properties |
|---|
| General Function | positive regulation of transcription from RNA polymerase II promoter in response to osmotic stress |
|---|
| Specific Function | Binds to the CRE motif 5'-TGACGTCA-3' and acts as a repressor of transcription of the SUC2 gene and most probably other genes. |
|---|
| Cellular Location | Not Available |
|---|
| SMPDB Pathways | | Stress-activated signalling pathways: high osmolarity test 1 | PW002773 | 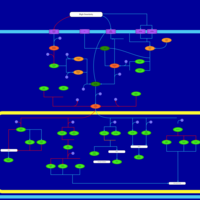 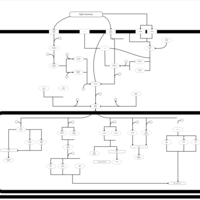 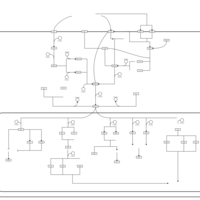 |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Metabolites | |
|---|
| GO Classification | | Function |
|---|
| transcriptional repressor activity, RNA polymerase II transcription factor binding | | positive regulation of transcription from RNA polymerase II promoter in response to osmotic stress | | mitogen-activated protein kinase binding | | cytosol | | nucleus | | RNA polymerase II core promoter proximal region sequence-specific DNA binding | | RNA polymerase II repressing transcription factor binding | | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding | | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding | | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding | | negative regulation of transcription from RNA polymerase II promoter |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | Not Available |
|---|
| Locus | |
|---|
| Gene Sequence | >1944 bp
ATGTCAAGCGAGGAACGCTCGAGACAACCAAGTACTGTTTCAACTTTCGATTTAGAACCC
AATCCTTTTGAACAAAGCTTCGCCTCTTCCAAGAAGGCTTTGTCACTTCCAGGCACGATC
TCCCATCCGTCTCTACCAAAAGAGCTTTCACGAAACAATTCTACCTCAACGATAACACAA
CATTCCCAACGTTCCACTCATAGTTTGAACAGTATTCCAGAAGAAAATGGGAATTCAACC
GTTACAGATAATAGTAATCATAATGACGTAAAAAAAGACTCACCTAGTTTTTTACCAGGC
CAACAAAGACCTACTATAATATCTCCGCCTATTCTCACACCTGGTGGGTCGAAAAGATTA
CCACCTCTACTCCTTTCTCCCTCTATTTTATATCAGGCAAATTCAACTACGAATCCCAGT
CAGAATTCACATTCTGTTTCAGTCTCTAATTCAAACCCTAGTGCAATAGGCGTTTCTTCT
ACCTCCGGATCTCTGTATCCAAACAGCTCTTCTCCTTCAGGGACTTCACTTATACGCCAG
CCACGAAACTCGAACGTGACCACAAGTAATTCTGGTAACGGCTTCCCCACGAATGATTCA
CAAATGCCTGGCTTTTTATTAAACTTGTCCAAATCTGGGTTGACACCTAATGAGTCCAAT
ATCAGGACCGGGTTGACACCCGGTATTCTTACGCAATCTTACAATTATCCCGTATTGCCG
TCAATTAATAAAAATACTATAACAGGTAGCAAAAATGTCAACAAAAGTGTCACAGTGAAT
GGAAGTATTGAAAACCACCCTCATGTTAATATAATGCACCCAACTGTAAATGGTACACCA
CTTACGCCGGGATTGAGTTCTCTGCTAAACTTACCATCTACTGGAGTTTTGGCTAATCCA
GTATTCAAATCAACACCTACAACAAATACCACAGATGGTACCGTCAACAACAGCATCAGT
AATTCCAATTTTTCCCCAAATACTTCAACGAAAGCGGCTGTCAAAATGGATAATCCGGCA
GAGTTCAATGCCATCGAGCACTCCGCTCATAATCACAAGGAGAATGAAAATTTAACGACT
CAAATTGAGAACAATGACCAGTTCAATAACAAAACACGAAAAAGAAAGAGAAGGATGTCT
AGCACAAGTTCTACTTCTAAGGCTTCAAGAAAAAATTCCATATCAAGAAAAAACTCAGCA
GTTACGACTGCACCAGCACAAAAAGATGATGTTGAAAATAATAAAATTTCAAACAACGTA
ACACTTGATGAGAATGAAGAGCAAGAAAGAAAAAGAAAGGAGTTTTTAGAAAGAAATAGA
GTAGCTGCATCTAAATTTAGAAAAAGAAAGAAAGAGTACATCAAGAAAATTGAAAATGAT
CTACAATTCTATGAATCTGAATATGACGATCTAACTCAGGTCATTGGGAAGCTATGCGGC
ATAATACCCTCAAGCTCCTCGAATTCCCAATTCAATGTGAATGTTTCAACTCCGTCATCA
TCATCACCACCATCTACATCTTTAATAGCATTGTTAGAGAGTAGCATTTCAAGGAGTGAT
TATTCAAGTGCAATGTCAGTATTATCAAACATGAAGCAATTGATATGTGAAACGAATTTT
TACCGAAGGGGAGGCAAAAACCCAAGGGACGACATGGATGGCCAAGAAGACAGCTTCAAT
AAGGACACTAACGTTGTCAAAAGCGAAAATGCGGGCTATCCTTCCGTTAATTCAAGACCA
ATAATTCTAGATAAAAAATACTCACTGAACTCTGGAGCAAATATCAGCAAAAGTAACACA
ACTACTAATAATGTGGGAAATAGTGCACAGAATATAATCAATTCATGCTACTCTGTTACT
AATCCATTGGTAATAAATGCAAATTCCGATACCCATGATACTAATAAGCATGATGTACTA
TCCACTCTACCTCACAATAATTGA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | Not Available |
|---|
| Protein Residues | 647 |
|---|
| Protein Molecular Weight | 70191.49 |
|---|
| Protein Theoretical pI | Not Available |
|---|
| Signalling Regions | Not Available |
|---|
| Transmembrane Regions | Not Available |
|---|
| Protein Sequence | >CRE-binding bZIP protein SKO1
MSSEERSRQPSTVSTFDLEPNPFEQSFASSKKALSLPGTISHPSLPKELSRNNSTSTITQ
HSQRSTHSLNSIPEENGNSTVTDNSNHNDVKKDSPSFLPGQQRPTIISPPILTPGGSKRL
PPLLLSPSILYQANSTTNPSQNSHSVSVSNSNPSAIGVSSTSGSLYPNSSSPSGTSLIRQ
PRNSNVTTSNSGNGFPTNDSQMPGFLLNLSKSGLTPNESNIRTGLTPGILTQSYNYPVLP
SINKNTITGSKNVNKSVTVNGSIENHPHVNIMHPTVNGTPLTPGLSSLLNLPSTGVLANP
VFKSTPTTNTTDGTVNNSISNSNFSPNTSTKAAVKMDNPAEFNAIEHSAHNHKENENLTT
QIENNDQFNNKTRKRKRRMSSTSSTSKASRKNSISRKNSAVTTAPAQKDDVENNKISNNV
TLDENEEQERKRKEFLERNRVAASKFRKRKKEYIKKIENDLQFYESEYDDLTQVIGKLCG
IIPSSSSNSQFNVNVSTPSSSSPPSTSLIALLESSISRSDYSSAMSVLSNMKQLICETNF
YRRGGKNPRDDMDGQEDSFNKDTNVVKSENAGYPSVNSRPIILDKKYSLNSGANISKSNT
TTNNVGNSAQNIINSCYSVTNPLVINANSDTHDTNKHDVLSTLPHNN |
|---|
| References |
|---|
| External Links | | Resource | Link |
|---|
| Saccharomyces Genome Database | SKO1 | | Uniprot ID | Q02100 | | Uniprot Name | SKO1 |
|
|---|
| General Reference | Not Available |
|---|