| Identification |
|---|
| Name | GTPase-activating protein BEM3 |
|---|
| Synonyms | |
|---|
| Gene Name | BEM3 |
|---|
| Enzyme Class | Not Available |
|---|
| Biological Properties |
|---|
| General Function | signal transduction |
|---|
| Specific Function | GTPase-activating protein (GAP) for CDC42 and less efficiently for RHO1. Negative regulator of the pheromone-response pathway through the STE20 protein kinase. |
|---|
| Cellular Location | Not Available |
|---|
| SMPDB Pathways | | Stress-activated signalling pathways: pheromone stress test 1 | PW012843 |  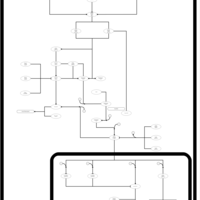 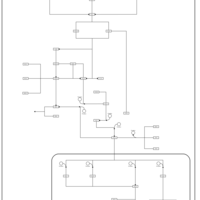 |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Metabolites | |
|---|
| GO Classification | | Function |
|---|
| cytoplasm | | cellular bud tip | | incipient cellular bud site | | mating projection tip | | septin ring organization | | cell cortex | | GTPase activator activity | | signal transduction | | negative regulation of Rho protein signal transduction | | establishment of cell polarity | | phosphatidylinositol-3-phosphate binding |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | Not Available |
|---|
| Locus | |
|---|
| Gene Sequence | >3387 bp
ATGACAGATAATTTGACCACAACTCATGGTGGTAGTACCACCCTTGAATTGTTGGCGCAG
TACAATGATCATAGATCCAAAAAGGACAAATCTATAGAACATATAGAAAAAGGAACATGT
TCAGGAAAAGAAAGAAATCCGTCCTATGATGAAATTTTTACGGAAAACATTAAGCTAAAG
CTACAAGTCCAAGAATATGAAACTGAAATTGAGAGTTTGGAAAAGGTTATTGATATGTTA
CAAAAAAACAGAGAAGCTTCCTTAGAAGTTGTGCTTGAGCAAGTTCAAAATGACTCAAGA
GATTCATACGTTAACGATCAATCATTTGTGTTACCCCCAAGATCTGCTGAAAGGAAAGCG
CATATCAAGAGTCTTAATTTGCCAATACCGACATTGTCCCCTCCATTGCAACAAGGTTCA
GACGTTGCACTTGAAACAAGTGTAACGCCTACAGTACCTCAAATAGGTGTTACATCAAAT
ACTTCAATAAGTAGGAAGCATTTACAGAACATGATACTAAACGATGAAATAGAAGCAAAT
AGTAGCTTCTCATCTCCCAAAATTATCAATAGATCTGTCTCAAGCCCTACAAAAATACAT
TCAGAACAGTTGGCTAGCCCAGCTGCCTCTGTTACCTATACGACGTCTAGGATAACCATA
AAATCACCTAATAAAGGTTCGAAATCTCCATTGCAAGAACGTTTGCGTTCCCCTCAAAAT
CCTAACAGAATGACGGCTGTTATCAACAACCATTTGCATTCTCCATTAAAGGCCAGTACC
TCTAATAATTTAGATGAGCTGACTGAAAGTAAAAGTCAACAACTAACTAATGACGCGATA
CAAAAAAATGACCGCGTTTATTCTTCTATAACATCCTCTGCATACACTACTGGAACTCCT
ACTTCAGCAGCCAAAAGTCCATCATCCCTTTTGGAAGTCAAGGAGGGTGAAAACAAGGCT
TTGGGTTTCTCTCCAGCATCTAAAGAAAAACTTGACGACTTCACCCAATTACTTGACTCC
TCATTTGGCGAAGAGGATCTAGTCAATACGGACAGTAAAGATCCTCTGTCCATAAAATCC
ACGATTAACGAATCTTTACCCCCACCTCCAGCGCCTCCAACTTTTTTCTCACCTACTTCT
AGTGGAAATATAAAAAACTCTACTCCTTTATCAAGCCATTTGGCTTCTCCGGTTATCTTG
AATAAAAAGGATGATAATTTTGGTGCTCAAAGTGCAAAAAATTTAAAGAAACCTGTGTTA
ACTTCATCACTCCCCAATCTTTCTACAAAATTGTCAACAACAAGTCAAAATGCTTCTCTT
CCTCCAAATCCTCCCGTGGAAAGTTCATCAAAACAAAAACAACTTGGAGAAACGGCATCC
ATCCATTCAACCAATACCTTGAATACATTTAGCTCAACACCCCAAGGTTCGTTGAAGACA
CTTAGAAGACCTCATGCAAGTAGCGTGAGCACCGTGAAGTCTGTGGCCCAAAGTCTTAAG
TCGGATATTCCATTATTCGTTCAACCGGAGGATTTTGGCACTATCCAGATTGAAGTACTG
AGCACATTGTACAGGGATAATGAAGATGATTTATCCATTTTAATTGCAATCATAGACAGG
AAATCCGGTAAGGAGATGTTTAAATTTTCAAAATCTATTCATAAAGTTCGCGAATTAGAT
GTATATATGAAATCACATGTTCCCGATTTACCATTGCCCACGTTACCTGATAGACAATTA
TTTCAGACTTTATCACCTACAAAAGTGGATACGCGAAAGAACATATTGAATCAGTATTAT
ACAAGCATATTCAGTGTCCCTGAATTTCCCAAAAATGTTGGTTTAAAAATCGCCCAATTT
ATTAGTACAGATACAGTAATGACACCTCCAATGATGGATGATAATGTGAAAGACGGATCA
CTTTTGCTGAGAAGACCAAAGACTCTAACAGGTAATAGTACATGGAGAGTTCGGTACGGT
ATTTTGCGCGATGATGTTTTACAATTGTTTGACAAAAATCAGTTGACTGAAACTATTAAA
TTGAGACAATCTTCTATAGAACTTATACCCAATCTCCCTGAAGATAGATTTGGAACGAGA
AATGGGTTTTTGATCACTGAGCATAAGAAAAGCGGACTGTCAACCTCAACAAAGTATTAT
ATTTGTACTGAAACGTCGAAGGAACGTGAATTATGGCTTTCTGCGTTTAGTGATTACATT
GATCCTTCACAAAGTTTATCGTTATCTAGCTCGAGGAATGCAAACGATACTGATTCTGCA
TCTCACTTGAGCGCAGGAACACACCATTCTAAATTTGGAAACGCAACTATCTCAGCAACC
GATACTCCATCATATGTCACTGATTTGACCCAAGAATATAATAATAATAACAATATTAGT
AATAGTAGCAATAACATTGCTAATAGCGATGGTATTGACTCAAACCCATCTTCTCACTCT
AACTTCCTAGCAAGTTCAAGCGGGAATGCAGAGGAGGAGAAGGATAGTAGGAGAGCCAAA
ATGAGAAGTTTATTCCCTTTTAAAAAATTAACCGGCCCGGCGAGTGCAATGAACCATATA
GGTATTACAATTTCAAATGATTCGGATTCTCCAACATCTCCTGACTCTATCATTAAGTCA
CCAAGCAAAAAATTAATGGAAGTATCCTCTTCGTCAAATTCTTCTACGGGTCCACATGTG
TCCACTGCAATATTTGGCTCGTCACTAGAAACGTGTTTAAGATTGAGTTCGCATAAGTAC
CAAAACGTTTACGATTTACCGAGTGTAGTTTATCGTTGCTTAGAGTATCTTTACAAAAAC
CGTGGTATCCAGGAAGAAGGCATATTTAGACTAAGCGGGTCCAGTACAGTTATCAAAACT
TTACAAGAAAGGTTTGATAAAGAGTACGATGTAGATTTATGTCGGTACAATGAAAGTATT
GAAGCTAAAGATGATGAGGCATCTCCAAGTCTTTACATTGGTGTGAATACTGTAAGTGGG
TTACTAAAGTTGTACTTGAGAAAGTTGCCCCATTTATTATTTGGTGACGAACAATTTTTG
TCGTTTAAAAGAGTAGTTGATGAAAATCATAATAATCCTGTTCAAATATCATTGGGGTTT
AAGGAACTTATTGAATCTGGCCTAGTTCCTCACGCTAACCTATCGTTGATGTATGCTCTT
TTTGAACTGCTGGTAAGGATCAATGAGAATAGTAAGTTTAATAAGATGAATCTAAGAAAT
TTATGCATCGTGTTTTCCCCAACTCTAAATATTCCAATTAGTATGCTTCAACCATTCATC
ACTGACTTTGCGTGTATATTTCAGGGTGGTGAACCAGTTAAAGAAGAGGAAAGGGAGAAA
GTAGATATACATATTCCTCAGGTTTGA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | Not Available |
|---|
| Protein Residues | 1128 |
|---|
| Protein Molecular Weight | 124911.845 |
|---|
| Protein Theoretical pI | Not Available |
|---|
| Signalling Regions | Not Available |
|---|
| Transmembrane Regions | Not Available |
|---|
| Protein Sequence | >GTPase-activating protein BEM3
MTDNLTTTHGGSTTLELLAQYNDHRSKKDKSIEHIEKGTCSGKERNPSYDEIFTENIKLK
LQVQEYETEIESLEKVIDMLQKNREASLEVVLEQVQNDSRDSYVNDQSFVLPPRSAERKA
HIKSLNLPIPTLSPPLQQGSDVALETSVTPTVPQIGVTSNTSISRKHLQNMILNDEIEAN
SSFSSPKIINRSVSSPTKIHSEQLASPAASVTYTTSRITIKSPNKGSKSPLQERLRSPQN
PNRMTAVINNHLHSPLKASTSNNLDELTESKSQQLTNDAIQKNDRVYSSITSSAYTTGTP
TSAAKSPSSLLEVKEGENKALGFSPASKEKLDDFTQLLDSSFGEEDLVNTDSKDPLSIKS
TINESLPPPPAPPTFFSPTSSGNIKNSTPLSSHLASPVILNKKDDNFGAQSAKNLKKPVL
TSSLPNLSTKLSTTSQNASLPPNPPVESSSKQKQLGETASIHSTNTLNTFSSTPQGSLKT
LRRPHASSVSTVKSVAQSLKSDIPLFVQPEDFGTIQIEVLSTLYRDNEDDLSILIAIIDR
KSGKEMFKFSKSIHKVRELDVYMKSHVPDLPLPTLPDRQLFQTLSPTKVDTRKNILNQYY
TSIFSVPEFPKNVGLKIAQFISTDTVMTPPMMDDNVKDGSLLLRRPKTLTGNSTWRVRYG
ILRDDVLQLFDKNQLTETIKLRQSSIELIPNLPEDRFGTRNGFLITEHKKSGLSTSTKYY
ICTETSKERELWLSAFSDYIDPSQSLSLSSSRNANDTDSASHLSAGTHHSKFGNATISAT
DTPSYVTDLTQEYNNNNNISNSSNNIANSDGIDSNPSSHSNFLASSSGNAEEEKDSRRAK
MRSLFPFKKLTGPASAMNHIGITISNDSDSPTSPDSIIKSPSKKLMEVSSSSNSSTGPHV
STAIFGSSLETCLRLSSHKYQNVYDLPSVVYRCLEYLYKNRGIQEEGIFRLSGSSTVIKT
LQERFDKEYDVDLCRYNESIEAKDDEASPSLYIGVNTVSGLLKLYLRKLPHLLFGDEQFL
SFKRVVDENHNNPVQISLGFKELIESGLVPHANLSLMYALFELLVRINENSKFNKMNLRN
LCIVFSPTLNIPISMLQPFITDFACIFQGGEPVKEEEREKVDIHIPQV |
|---|
| References |
|---|
| External Links | | Resource | Link |
|---|
| Saccharomyces Genome Database | BEM3 | | Uniprot ID | P32873 | | Uniprot Name | BEM3 |
|
|---|
| General Reference | Not Available |
|---|