| Identification |
|---|
| Name | Cell division control protein 42 |
|---|
| Synonyms | - Suppressor of RHO3 protein 2
|
|---|
| Gene Name | CDC42 |
|---|
| Enzyme Class | Not Available |
|---|
| Biological Properties |
|---|
| General Function | small GTPase mediated signal transduction |
|---|
| Specific Function | Involved in development of cell polarity during the cell division cycle, and essential for bud emergence. Affects signaling in the pheromone-response pathway through the STE20 protein kinase. Negatively regulated by the GTPase-activating proteins RGA1, BEM3, and BEM4. |
|---|
| Cellular Location | Not Available |
|---|
| SMPDB Pathways | | Stress-activated signalling pathways: high osmolarity 1469654356 | PW002970 |    | | Stress-activated signalling pathways: high osmolarity test 1 | PW002773 | 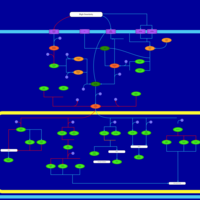 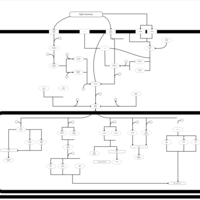 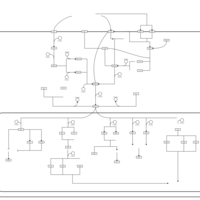 | | Stress-activated signalling pathways: pheromone stress test 1 | PW012843 |  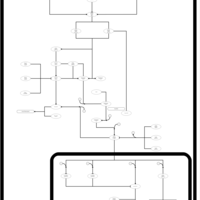 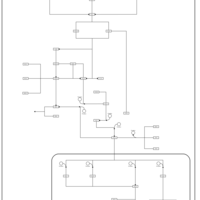 | | Stress-activated signalling pathways: starvation test 1 | PW003483 | 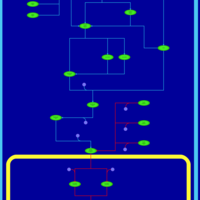 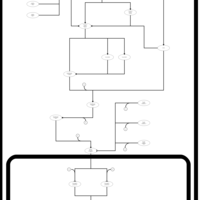 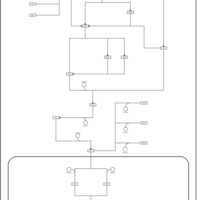 |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Metabolites | |
|---|
| GO Classification | | Function |
|---|
| small GTPase mediated signal transduction | | regulation of initiation of mating projection growth | | invasive growth in response to glucose limitation | | plasma membrane | | regulation of exit from mitosis | | septin ring organization | | conjugation with cellular fusion | | cell cycle | | pheromone-dependent signal transduction involved in conjugation with cellular fusion | | positive regulation of pseudohyphal growth | | cellular bud neck | | fungal-type vacuole membrane | | GTP binding | | nuclear membrane | | GTPase activity | | septin ring | | cellular bud tip | | budding cell apical bud growth | | incipient cellular bud site | | budding cell isotropic bud growth | | mating projection tip | | establishment of cell polarity | | regulation of exocyst localization | | positive regulation of exocytosis | | regulation of vacuole fusion, non-autophagic | | regulation of cell morphogenesis |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | Not Available |
|---|
| Locus | |
|---|
| Gene Sequence | >576 bp
ATGCAAACGCTAAAGTGTGTTGTTGTCGGTGATGGTGCTGTTGGGAAAACGTGCCTTCTA
ATCTCCTATACAACGAATCAATTTCCAGCCGACTATGTTCCAACAGTGTTCGATAACTAT
GCGGTGACTGTGATGATTGGTGATGAACCATATACGTTAGGTTTGTTTGATACGGCCGGT
CAAGAAGATTACGATCGATTGAGACCCTTGTCATATCCTTCTACTGATGTATTTTTGGTT
TGTTTCAGTGTTATTTCCCCACCCTCTTTTGAAAACGTTAAAGAAAAATGGTTCCCTGAA
GTACATCACCATTGTCCAGGTGTACCATGCCTGGTCGTCGGTACGCAGATTGATCTAAGG
GATGACAAGGTAATCATCGAGAAGTTGCAAAGACAAAGATTACGTCCGATTACATCAGAA
CAAGGTTCCAGGTTAGCAAGAGAACTGAAAGCAGTAAAATATGTCGAGTGTTCGGCACTA
ACACAACGCGGTTTGAAGAATGTATTCGATGAAGCTATCGTGGCCGCCTTGGAGCCTCCT
GTTATCAAGAAAAGTAAAAAATGTGCAATTTTGTAG |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | Not Available |
|---|
| Protein Residues | 191 |
|---|
| Protein Molecular Weight | 21322.54 |
|---|
| Protein Theoretical pI | Not Available |
|---|
| Signalling Regions | Not Available |
|---|
| Transmembrane Regions | Not Available |
|---|
| Protein Sequence | >Cell division control protein 42
MQTLKCVVVGDGAVGKTCLLISYTTNQFPADYVPTVFDNYAVTVMIGDEPYTLGLFDTAG
QEDYDRLRPLSYPSTDVFLVCFSVISPPSFENVKEKWFPEVHHHCPGVPCLVVGTQIDLR
DDKVIIEKLQRQRLRPITSEQGSRLARELKAVKYVECSALTQRGLKNVFDEAIVAALEPP
VIKKSKKCAIL |
|---|
| References |
|---|
| External Links | | Resource | Link |
|---|
| Saccharomyces Genome Database | CDC42 | | Uniprot ID | P19073 | | Uniprot Name | CDC42 |
|
|---|
| General Reference | Not Available |
|---|