| Identification |
|---|
| Name | MAP kinase kinase PBS2 |
|---|
| Synonyms | - Polymyxin B resistance protein 2
- Suppressor of fluoride sensitivity 4
|
|---|
| Gene Name | PBS2 |
|---|
| Enzyme Class | Not Available |
|---|
| Biological Properties |
|---|
| General Function | response to antibiotic |
|---|
| Specific Function | Kinase involved in a signal transduction pathway that is activated by changes in the osmolarity of the extracellular environment. Seems to phosphorylate HOG1 on a tyrosine residue. |
|---|
| Cellular Location | Not Available |
|---|
| SMPDB Pathways | | Stress-activated signalling pathways: high osmolarity | PW002516 | 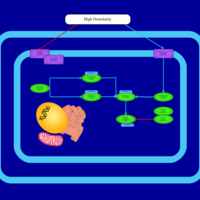 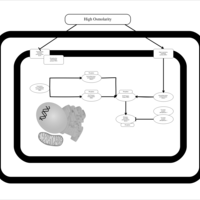 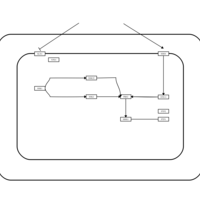 | | Stress-activated signalling pathways: high osmolarity 1469654356 | PW002970 |    |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Metabolites | |
|---|
| GO Classification | | Function |
|---|
| protein tyrosine kinase activity | | actin filament organization | | activation of MAPK activity involved in osmosensory signaling pathway | | cellular response to heat | | hyperosmotic response | | MAPK import into nucleus involved in osmosensory signaling pathway | | mitotic cell cycle | | ATP binding | | N-terminal peptidyl-methionine acetylation | | cellular bud neck | | osmosensory signaling pathway | | cytoplasm | | response to antibiotic | | cellular bud tip | | MAP kinase kinase activity | | receptor signaling protein serine/threonine kinase activity | | protein phosphorylation | | NatB complex | | MAP-kinase scaffold activity | | peptide alpha-N-acetyltransferase activity |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | Not Available |
|---|
| Locus | |
|---|
| Gene Sequence | >2007 bp
ATGGAAGACAAGTTTGCTAACCTCAGTCTCCATGAGAAAACTGGTAAGTCATCTATCCAA
TTAAACGAGCAAACAGGCTCAGATAATGGCTCTGCTGTCAAGAGAACATCTTCGACGTCC
TCGCACTACAATAACATCAACGCTGACCTTCATGCTCGTGTAAAAGCTTTTCAAGAACAA
CGTGCATTGAAAAGGTCTGCCAGCGTGGGCAGTAATCAAAGCGAGCAAGACAAAGGCAGT
TCACAATCACCTAAACATATTCAGCAGATTGTTAATAAGCCATTGCCGCCTCTTCCCGTA
GCAGGAAGTTCTAAGGTTTCACAAAGAATGAGTAGCCAAGTCGTGCAAGCGTCCTCCAAG
AGCACTCTTAAGAACGTTCTGGACAATCAAGAAACACAAAACATTACCGACGTAAATATT
AACATCGATACAACCAAAATTACCGCCACAACAATTGGTGTAAATACTGGCCTACCTGCT
ACTGACATTACGCCGTCAGTTTCTAATACTGCATCAGCAACACATAAGGCGCAATTGCTG
AATCCTAACAGAAGGGCACCAAGAAGGCCGCTTTCTACCCAGCACCCTACAAGACCAAAT
GTTGCCCCGCATAAGGCCCCTGCTATAATCAACACACCAAAACAAAGTTTAAGTGCCCGT
CGAGGGCTCAAATTACCACCAGGAGGAATGTCATTAAAAATGCCCACTAAAACAGCTCAA
CAGCCGCAGCAGTTTGCCCCAAGCCCTTCAAACAAAAAACATATAGAAACCTTATCAAAC
AGCAAAGTTGTTGAAGGGAAAAGATCGAATCCGGGTTCTTTGATAAATGGTGTGCAAAGC
ACATCCACCTCATCAAGTACCGAAGGCCCACATGACACTGTAGGCACTACACCCAGAACT
GGAAACAGCAACAACTCTTCAAATTCTGGTAGTAGTGGTGGTGGTGGTCTTTTCGCAAAT
TTCTCGAAATACGTGGATATCAAATCCGGCTCTTTGAATTTTGCAGGCAAACTATCGCTA
TCCTCTAAAGGAATAGATTTCAGCAATGGTTCTAGTTCGAGAATTACATTGGACGAACTA
GAATTTTTGGATGAACTGGGTCATGGTAACTATGGTAACGTCTCAAAGGTACTGCATAAG
CCCACAAATGTTATTATGGCGACGAAGGAAGTCCGTTTGGAGCTAGATGAGGCTAAATTT
AGACAAATTTTAATGGAACTAGAAGTTTTGCATAAATGCAATTCTCCCTATATTGTGGAT
TTTTATGGTGCATTCTTTATTGAGGGCGCCGTCTACATGTGTATGGAATACATGGATGGT
GGTTCCTTGGATAAAATATACGACGAATCATCTGAAATCGGCGGCATTGATGAACCTCAG
CTAGCGTTTATTGCCAATGCTGTCATTCATGGACTAAAAGAACTCAAAGAGCAGCATAAT
ATCATACACAGAGATGTCAAACCAACAAATATTTTATGTTCAGCCAACCAAGGCACCGTA
AAGCTGTGCGATTTCGGTGTTTCTGGTAATTTGGTGGCATCTTTAGCGAAGACTAATATT
GGTTGTCAGTCATACATGGCACCTGAACGAATCAAATCGTTGAATCCAGATAGAGCCACC
TATACCGTACAGTCAGACATCTGGTCTTTAGGTTTAAGCATTCTGGAAATGGCACTAGGT
AGATATCCGTATCCACCAGAAACATACGACAACATTTTCTCTCAATTGAGCGCTATTGTT
GATGGGCCGCCACCGAGATTACCTTCAGATAAATTCAGTTCTGACGCACAAGATTTTGTT
TCTTTATGTCTACAAAAGATTCCGGAAAGAAGACCTACATACGCAGCTTTAACAGAGCAT
CCTTGGTTAGTAAAATACAGAAACCAGGATGTCCACATGAGTGAGTATATCACTGAACGA
TTAGAAAGGCGCAACAAAATCTTACGGGAACGTGGTGAGAATGGTTTATCTAAAAATGTA
CCGGCATTACATATGGGTGGTTTATAG |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | Not Available |
|---|
| Protein Residues | 668 |
|---|
| Protein Molecular Weight | 72719.245 |
|---|
| Protein Theoretical pI | Not Available |
|---|
| PDB File | show |
|---|
| Signalling Regions | Not Available |
|---|
| Transmembrane Regions | Not Available |
|---|
| Protein Sequence | >MAP kinase kinase PBS2
MEDKFANLSLHEKTGKSSIQLNEQTGSDNGSAVKRTSSTSSHYNNINADLHARVKAFQEQ
RALKRSASVGSNQSEQDKGSSQSPKHIQQIVNKPLPPLPVAGSSKVSQRMSSQVVQASSK
STLKNVLDNQETQNITDVNINIDTTKITATTIGVNTGLPATDITPSVSNTASATHKAQLL
NPNRRAPRRPLSTQHPTRPNVAPHKAPAIINTPKQSLSARRGLKLPPGGMSLKMPTKTAQ
QPQQFAPSPSNKKHIETLSNSKVVEGKRSNPGSLINGVQSTSTSSSTEGPHDTVGTTPRT
GNSNNSSNSGSSGGGGLFANFSKYVDIKSGSLNFAGKLSLSSKGIDFSNGSSSRITLDEL
EFLDELGHGNYGNVSKVLHKPTNVIMATKEVRLELDEAKFRQILMELEVLHKCNSPYIVD
FYGAFFIEGAVYMCMEYMDGGSLDKIYDESSEIGGIDEPQLAFIANAVIHGLKELKEQHN
IIHRDVKPTNILCSANQGTVKLCDFGVSGNLVASLAKTNIGCQSYMAPERIKSLNPDRAT
YTVQSDIWSLGLSILEMALGRYPYPPETYDNIFSQLSAIVDGPPPRLPSDKFSSDAQDFV
SLCLQKIPERRPTYAALTEHPWLVKYRNQDVHMSEYITERLERRNKILRERGENGLSKNV
PALHMGGL |
|---|
| References |
|---|
| External Links | | Resource | Link |
|---|
| Saccharomyces Genome Database | PBS2 | | Uniprot ID | P08018 | | Uniprot Name | PBS2 | | PDB ID | 2VKN |
|
|---|
| General Reference | Not Available |
|---|