| Identification |
|---|
| Name | Putative inosine-5'-monophosphate dehydrogenase IMD1 |
|---|
| Synonyms | - IMP dehydrogenase
- IMPD
- IMPDH
|
|---|
| Gene Name | IMD1 |
|---|
| Enzyme Class | |
|---|
| Biological Properties |
|---|
| General Function | Involved in catalytic activity |
|---|
| Specific Function | Inosine 5'-phosphate + NAD(+) + H(2)O = xanthosine 5'-phosphate + NADH |
|---|
| Cellular Location | Not Available |
|---|
| SMPDB Pathways | | purine nucleotides de novo biosynthesis | PW002478 |  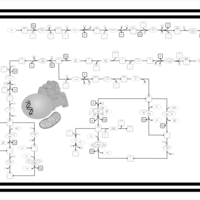 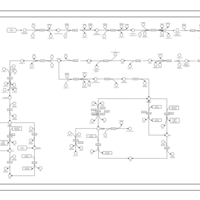 |
|
|---|
| KEGG Pathways | |
|---|
| SMPDB Reactions | |
|---|
| KEGG Reactions | Not Available |
|---|
| Metabolites | | YMDB ID | Name | View |
|---|
| YMDB00110 | NAD | Show | | YMDB00143 | NADH | Show | | YMDB00289 | Xanthylic acid | Show | | YMDB00352 | Inosinic acid | Show | | YMDB00862 | hydron | Show | | YMDB00890 | water | Show |
|
|---|
| GO Classification | | Component |
|---|
| Not Available | | Function |
|---|
| catalytic activity | | oxidoreductase activity | | Process |
|---|
| metabolic process | | oxidation reduction |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | chromosome 1 |
|---|
| Locus | YAR073W |
|---|
| Gene Sequence | >1212 bp
ATGGCCGCCATTAGAGACTACAAGACCGCACTAGATCTTACCAAGAGCCTACCAAGACCG
GATGGTTTGTCAGTGCAGGAACTGATGGACTCCAAGATCAGAGGTGGGTTGGCTTATAAC
GATTTTTTAATCTTACCAGGTTTAGTCGATTTTGCGTCCTCTGAAGTTAGCCTACAGACC
AAGCTAACCAGGAATATTACTTTAAACATTCCATTAGTATCCTCTCCAATGGACACTGTG
ACGGAATCTGAAATGGCCACTTTTATGGCTCTGTTGGATGGTATCGGTTTCATTCACCAT
AACTGTACTCCAGAGGACCAAGCTGACATGGTCAGAAGAGTCAAGAACTATGAAAATGGG
TTTATTAACAACCCTATAGTGATTTCTCCAACTACGACCGTTGGTGAAGCTAAGAGCATG
AAGGAAAAGTATGGATTTGCAGGCTTCCCTGTCACGGCAGATGGAAAGAGAAATGCAAAG
TTGGTGGGTGCCATCACCTCTCGTGATATACAATTCGTTGAGGACAACTCTTTACTCGTT
CAGGATGTCATGACCAAAAACCCTGTTACCGGCGCACAAGGTATCACATTATCAGAAGGT
AACGAAATTCTAAAGAAAATCAAAAAGGGTAGGCTACTGGTTGTTGATGAAAAGGGTAAC
TTAGTTTCTATGCTTTCCCGAACTGATTTAATGAAAAATCAGAAGTACCCATTAGCGTCC
AAATCTGCCAACACCAAGCAACTGTTATGGGGTGCTTCTATTGGGACTATGGACGCTGAT
AAAGAAAGACTAAGATTATTGGTAAAAGCTGGCTTGGATGTCGTCATATTGGATTCCTCT
CAAGGTAACTCTATTTTCCAATTGAACATGATCAAATGGATTAAAGAAACTTTCCCAGAT
TTGGAAATCATTGCTGGTAACGTTGTCACCAAGGAACAAGCTGCCAATTTGATTGCTGCC
GGTGCGGACGGTTTGAGAATTGGTATGGGAACTGGCTCTATTTGTATTACCCAAAAAGTT
ATGGCTTGTGGTAGGCCACAAGGTACAGCCGTCTACAACGTGTGTGAATTTGCTAACCAA
TTCGGTGTTCCATGTATGGCTGATGGTGGTGTTCAAAAACATTGGTCATATTATTACCAA
AGCTTTGGCTCTTGGTTCTTCTACTGTTATGATGGGTGGTATGTTGGCCGGTACTACCGA
ATCACCAGGTGA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | |
|---|
| Protein Residues | 403 |
|---|
| Protein Molecular Weight | 44385.80078 |
|---|
| Protein Theoretical pI | 8.32 |
|---|
| Signalling Regions | |
|---|
| Transmembrane Regions | |
|---|
| Protein Sequence | >Putative inosine-5'-monophosphate dehydrogenase IMD1
MAAIRDYKTALDLTKSLPRPDGLSVQELMDSKIRGGLAYNDFLILPGLVDFASSEVSLQT
KLTRNITLNIPLVSSPMDTVTESEMATFMALLDGIGFIHHNCTPEDQADMVRRVKNYENG
FINNPIVISPTTTVGEAKSMKEKYGFAGFPVTADGKRNAKLVGAITSRDIQFVEDNSLLV
QDVMTKNPVTGAQGITLSEGNEILKKIKKGRLLVVDEKGNLVSMLSRTDLMKNQKYPLAS
KSANTKQLLWGASIGTMDADKERLRLLVKAGLDVVILDSSQGNSIFQLNMIKWIKETFPD
LEIIAGNVVTKEQAANLIAAGADGLRIGMGTGSICITQKVMACGRPQGTAVYNVCEFANQ
FGVPCMADGGVQKHWSYYYQSFGSWFFYCYDGWYVGRYYRITR |
|---|
| References |
|---|
| External Links | |
|---|
| General Reference | - Bussey, H., Kaback, D. B., Zhong, W., Vo, D. T., Clark, M. W., Fortin, N., Hall, J., Ouellette, B. F., Keng, T., Barton, A. B., et, a. l. .. (1995). "The nucleotide sequence of chromosome I from Saccharomyces cerevisiae." Proc Natl Acad Sci U S A 92:3809-3813.7731988
- Hyle, J. W., Shaw, R. J., Reines, D. (2003). "Functional distinctions between IMP dehydrogenase genes in providing mycophenolate resistance and guanine prototrophy to yeast." J Biol Chem 278:28470-28478.12746440
- Barton, A. B., Kaback, D. B. (2006). "Telomeric silencing of an open reading frame in Saccharomyces cerevisiae." Genetics 173:1169-1173.16582424
- Albuquerque, C. P., Smolka, M. B., Payne, S. H., Bafna, V., Eng, J., Zhou, H. (2008). "A multidimensional chromatography technology for in-depth phosphoproteome analysis." Mol Cell Proteomics 7:1389-1396.18407956
|
|---|
