| Identification |
|---|
| Name | Protein SLG1 |
|---|
| Synonyms | - Cell wall integrity and stress response component 1
- Synthetic lethal with GAP protein 1
|
|---|
| Gene Name | SLG1 |
|---|
| Enzyme Class | Not Available |
|---|
| Biological Properties |
|---|
| General Function | Rho protein signal transduction |
|---|
| Specific Function | Plays a role during G1 to regulate entering or exiting the cell cycle. Involved in stress responses. Has a role in cell wall integrity signaling. Activates ROM1 or ROM2 catalyzed guanine nucleotide exchange toward RHO1. Important regulator of the actin cytoskeleton rearrangements in conditions of cell wall expansion and membrane stretching. Specifically required for the actin reorganization induced by hypo-osmotic shock. Multicopy suppressor of 1,3-beta-glucan synthase (GS). Activates GS upstream of RHO1. Acts positively on the PKC1-MAPK pathway. Activates transiently SLT2 during alkaline stress, which leads to an increase in the expression of several specific genes. |
|---|
| Cellular Location | Not Available |
|---|
| SMPDB Pathways | | Stress-activated signalling pathways: cell wall stress test 1 | PW007861 | 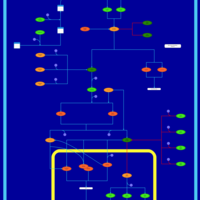 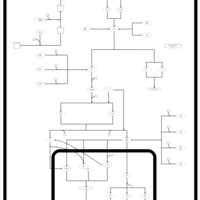 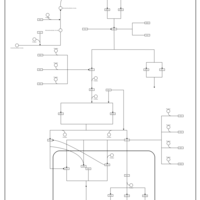 |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Metabolites | |
|---|
| GO Classification | | Function |
|---|
| transmembrane signaling receptor activity | | response to heat | | Rho protein signal transduction | | integral component of membrane | | cellular bud neck | | actin cytoskeleton organization | | positive regulation of endocytosis | | plasma membrane | | cell cycle | | establishment of cell polarity | | fungal-type cell wall organization | | pexophagy | | intracellular | | response to osmotic stress |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | Not Available |
|---|
| Locus | |
|---|
| Gene Sequence | >1137 bp
ATGAGACCGAACAAAACAAGTCTGCTTCTGGCGTTATTATCCATTTTATCGCAAGCGAAC
GCCTATGAATACGTGAATTGTTTTAGCTCACTACCCTCTGACTTTTCAAAGGCCGATTCA
TATAACTGGCAGTCGAGTTCACACTGTAACAGTGAGTGTAGCGCAAAAGGTGCAAGCTAC
TTTGCCCTTTATAATCATTCAGAATGTTATTGTGGTGATACTAATCCATCTGGTTCGGAA
TCTACTTCCTCTTCATGTAATACGTATTGCTTTGGTTACAGCAGTGAGATGTGTGGTGGT
GAAGATGCCTATTCTGTGTACCAACTTGACTCTGACACAAATAGCAATAGCATAAGCAGC
TCAGATTCAAGTACGGAGTCTACTTCTGCGTCGTCTTCCACAACTTCTTCAACAACGTCC
TCCACAACATCAACTACATCATCGACTACATCATCAACTACATCATCAATGGCGTCTTCC
TCTACAGTACAGAATTCCCCCGAGTCAACTCAAGCAGCTGCCTCTATTTCAACGTCACAG
AGTTCGAGCACTGTAACGTCAGAAAGCTCGCTAACTTCGGATACTTTGGCAACGAGCAGT
ACTAGCTCTCAATCACAGGACGCCACTTCGATAATCTATTCCACTACTTTTCACACTGAG
GGCGGTTCCACAATTTTTGTCACGAACACCATCACGGCAAGTGCACAGAATTCAGGATCG
GCCACAGGTACAGCTGGATCTGACTCTACGAGTGGAAGTAAAACTCATAAAAAGAAAGCC
AATGTAGGGGCAATTGTAGGCGGCGTTGTAGGTGGTGTAGTGGGAGCCGTAGCCATTGCT
CTTTGTATCTTGTTGATTGTCAGACACATTAATATGAAGCGGGAACAAGACAGGATGGAA
AAGGAATACCAAGAGGCGATAAAACCAGTTGAGTACCCTGATAAACTATACGCCTCTTCA
TTTTCATCTAATCACGGGCCCTCTTCAGGTAGCTTCGAGGAGGAGCACACCAAAGGGCAA
ACTGATATTAACCCTTTCGACGACTCAAGGAGAATAAGTAACGGAACATTCATAAATGGC
GGACCAGGAGGGAAAAACAACGTTTTAACAGTGGTCAATCCAGACGAAGCTGATTGA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | Not Available |
|---|
| Protein Residues | 378 |
|---|
| Protein Molecular Weight | 39269.585 |
|---|
| Protein Theoretical pI | Not Available |
|---|
| Signalling Regions | |
|---|
| Transmembrane Regions | |
|---|
| Protein Sequence | >Protein SLG1
MRPNKTSLLLALLSILSQANAYEYVNCFSSLPSDFSKADSYNWQSSSHCNSECSAKGASY
FALYNHSECYCGDTNPSGSESTSSSCNTYCFGYSSEMCGGEDAYSVYQLDSDTNSNSISS
SDSSTESTSASSSTTSSTTSSTTSTTSSTTSSTTSSMASSSTVQNSPESTQAAASISTSQ
SSSTVTSESSLTSDTLATSSTSSQSQDATSIIYSTTFHTEGGSTIFVTNTITASAQNSGS
ATGTAGSDSTSGSKTHKKKANVGAIVGGVVGGVVGAVAIALCILLIVRHINMKREQDRME
KEYQEAIKPVEYPDKLYASSFSSNHGPSSGSFEEEHTKGQTDINPFDDSRRISNGTFING
GPGGKNNVLTVVNPDEAD |
|---|
| References |
|---|
| External Links | | Resource | Link |
|---|
| Saccharomyces Genome Database | SLG1 | | Uniprot ID | P54867 | | Uniprot Name | SLG1 |
|
|---|
| General Reference | Not Available |
|---|