| Identification |
|---|
| Name | Protein MTL1 |
|---|
| Synonyms | |
|---|
| Gene Name | MTL1 |
|---|
| Enzyme Class | Not Available |
|---|
| Biological Properties |
|---|
| General Function | fungal-type cell wall organization |
|---|
| Specific Function | Involved in cell integrity signaling during vegetative growth at elevated temperature. Acts positively on the PKC1-MAPK pathway. Cell membrane sensor of oxidative stress in the cell integrity pathway upstream of PKC1. Required to transmit the oxidative signal to SLT2 and to restore the correct actin organization following oxidative stress. Multicopy suppressor of 1,3-beta-glucan synthase (GS) mutation. Also suppresses RGD1 null mutations. |
|---|
| Cellular Location | Not Available |
|---|
| SMPDB Pathways | | Stress-activated signalling pathways: cell wall stress test 1 | PW007861 | 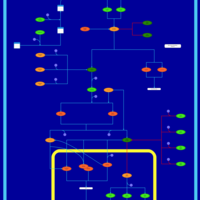 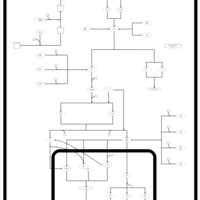 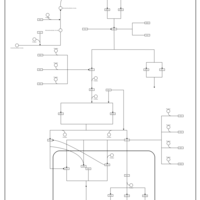 |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Metabolites | |
|---|
| GO Classification | | Function |
|---|
| integral component of plasma membrane | | cellular response to oxidative stress | | fungal-type cell wall organization | | cellular response to glucose starvation |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | Not Available |
|---|
| Locus | |
|---|
| Gene Sequence | >1656 bp
ATGGCAAGCTGCAATCCGACCAGGAAGAAGAGCTCTGCTTCAAGCCTATCTATGTGGAGA
ACGATTCTCATGGCGTTAACAACACTACCGCTAAGTGTTCTTTCGCAGGAGTTGGTTCCA
GCTAATAGCACAACATCGAGCACAGCTCCTTCCATCACTTCGCTTTCCGCAGTTGAGTCA
TTTACGTCCAGTACCGATGCAACGAGCAGCGCAAGTTTATCAACGCCGAGTATAGCTTCA
GTATCCTTTACTTCCTTCCCACAAAGTTCTTCACTGCTTACTCTTTCGTCAACATTATCC
TCAGAACTTTCCTCTTCGTCCATGCAAGTTTCGTCGTCTTCAACATCGTCGTCTTCTTCG
GAGGTTACGTCATCATCGTCATCATCATCAATATCTCCTTCCTCTTCATCATCAACAATA
ATATCATCGTCATCATCACTGCCGACATTCACTGTGGCATCAACATCTTCGACAGTTGCC
TCCTCCACACTTTCCACTAGCTCATCGTTGGTTATCTCTACGTCTTCGTCAACGTTTACG
TTTAGTTCGGAAAGTTCAAGCTCTTTGATTTCCTCTTCAATTTCAACATCCGTTTCGACT
TCTTCAGTGTACGTTCCCTCCTCTTCAACTTCATCTCCACCTTCGTCCTCATCCGAATTG
ACATCATCCTCGTACTCATCATCCTCATCCTCATCCACCCTCTTTTCCTACTCCTCCTCA
TTTTCATCATCCTCATCCTCATCATCCTCATCATCATCCTCATCCTCATCATCATCATCA
TCATCATCATCATATTTCACCCTCTCCACATCTTCCTCTTCATCCATATACTCGTCTTCG
TCATATCCTTCATTTTCATCTTCATCTTCCTCAAACCCTACCTCATCAATCACTTCTACA
TCCGCCTCATCTTCTATTACTCCCGCTTCCGAATATTCCAATTTGGCAAAAACCATAACT
AGTATAATAGAAGGCCAGACCATCCTCTCTAACTACTATACCACAATAACGTATTCACCG
ACAGCATCCGCATCTTCAGGAAAAAATTCACATCACTCAGGCTTATCAAAAAAGAATCGT
AATATTATCATCGGTTGTGTGGTTGGCATAGGTGCCCCCCTCATCCTAATTCTACTAATA
TTGATTTACATGTTTTGTGTTCAGCCTAAAAAAACGGATTTCATTGACTCTGACGGTAAA
ATTGTCACAGCTTATCGTAGTAACATTTTCACCAAAATATGGTATTTCTTGCTGGGTAAA
AAAATTGGTGAAACAGAAAGATTCAGCTCAGATTCCCCCATCGGCAGCAATAATATTCAG
AATTTTGGTGATATCGATCCAGAAGATATACTTAACAATGACAACCCCTACACCCCTAAA
CACACTAATGTTGAAGGCTACGACGACGACGACGACGACGACGCTAATGATGAAAACCTA
TCATCCAACTTCCATAACAGAGGCATAGATGATCAATACTCACCTACTAAATCTGCATCA
TATTCAATGTCGAATAGTAATAGTCAAGATTACAACGACGCAGATGAAGTAATGCACGAT
GAAAACATTCATCGTGTTTATGATGACAGCGAAGCTAGCATCGACGAGAACTATTACACG
AAACCAAACAACGGCTTAAATATCACGAACTATTAA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | Not Available |
|---|
| Protein Residues | 551 |
|---|
| Protein Molecular Weight | 57526.68 |
|---|
| Protein Theoretical pI | Not Available |
|---|
| Signalling Regions | |
|---|
| Transmembrane Regions | |
|---|
| Protein Sequence | >Protein MTL1
MASCNPTRKKSSASSLSMWRTILMALTTLPLSVLSQELVPANSTTSSTAPSITSLSAVES
FTSSTDATSSASLSTPSIASVSFTSFPQSSSLLTLSSTLSSELSSSSMQVSSSSTSSSSS
EVTSSSSSSSISPSSSSSTIISSSSSLPTFTVASTSSTVASSTLSTSSSLVISTSSSTFT
FSSESSSSLISSSISTSVSTSSVYVPSSSTSSPPSSSSELTSSSYSSSSSSSTLFSYSSS
FSSSSSSSSSSSSSSSSSSSSSSSYFTLSTSSSSSIYSSSSYPSFSSSSSSNPTSSITST
SASSSITPASEYSNLAKTITSIIEGQTILSNYYTTITYSPTASASSGKNSHHSGLSKKNR
NIIIGCVVGIGAPLILILLILIYMFCVQPKKTDFIDSDGKIVTAYRSNIFTKIWYFLLGK
KIGETERFSSDSPIGSNNIQNFGDIDPEDILNNDNPYTPKHTNVEGYDDDDDDDANDENL
SSNFHNRGIDDQYSPTKSASYSMSNSNSQDYNDADEVMHDENIHRVYDDSEASIDENYYT
KPNNGLNITNY |
|---|
| References |
|---|
| External Links | | Resource | Link |
|---|
| Saccharomyces Genome Database | MTL1 | | Uniprot ID | P53214 | | Uniprot Name | MTL1 |
|
|---|
| General Reference | Not Available |
|---|