| Identification |
|---|
| Name | RNA polymerase II-associated protein 1 |
|---|
| Synonyms | |
|---|
| Gene Name | PAF1 |
|---|
| Enzyme Class | Not Available |
|---|
| Biological Properties |
|---|
| General Function | transcription from RNA polymerase I promoter |
|---|
| Specific Function | The PAF1 complex is a multifunctional complex. Involved in transcription initiation via genetic interactions with TATA-binding proteins. Involved in elongation. It regulates 3'-end formation of snR47 by modulating the recruitment or stable association of NRD1 and NAB3 with RNA polymerase II. Also has a role in transcription-coupled histone modification. Required for activation of the RAD6/UBC2-BRE1 ubiquitin ligase complex, which ubiquitinates histone H2B to form H2BK123ub1. Also required for the methylation of histone H3 by the COMPASS complex to form H3K4me, by SET2 to form H3K36me, and by DOT1 to form H3K79me. RNA polymerase II associated protein important for transcription of a subset of genes. Required for both positive and negative regulation. Negatively regulates MAK16 expression. Also required for efficient CLN2 transcription in late G1 and may be involved in transcription of galactose-inducible genes. |
|---|
| Cellular Location | Not Available |
|---|
| SMPDB Pathways | | Stress-activated signalling pathways: cell wall stress test 1 | PW007861 | 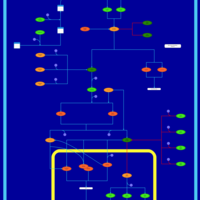 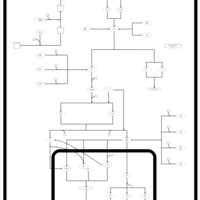 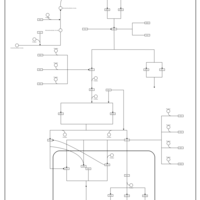 |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Metabolites | |
|---|
| GO Classification | | Function |
|---|
| negative regulation of transcription from RNA polymerase II promoter | | positive regulation of phosphorylation of RNA polymerase II C-terminal domain serine 2 residues | | chromatin binding | | positive regulation of transcription elongation from RNA polymerase I promoter | | positive regulation of transcription elongation from RNA polymerase II promoter | | regulation of chromatin silencing at telomere | | Cdc73/Paf1 complex | | regulation of histone H2B conserved C-terminal lysine ubiquitination | | transcriptionally active chromatin | | regulation of histone H2B ubiquitination | | RNA polymerase II C-terminal domain phosphoserine binding | | regulation of histone H3-K4 methylation | | transcription factor activity, RNA polymerase II core promoter sequence-specific | | regulation of phosphorylation of RNA polymerase II C-terminal domain serine 2 residues | | transcription factor activity, RNA polymerase II transcription factor binding | | regulation of transcription from RNA polymerase II promoter | | transcription factor activity, TFIIF-class transcription factor binding | | regulation of transcription involved in G1/S transition of mitotic cell cycle | | regulation of transcription-coupled nucleotide-excision repair | | chromatin silencing at rDNA | | rRNA processing | | DNA-templated transcription, termination | | snoRNA 3'-end processing | | global genome nucleotide-excision repair | | snoRNA transcription from an RNA polymerase II promoter | | histone modification | | transcription elongation from RNA polymerase I promoter | | mRNA 3'-end processing | | transcription elongation from RNA polymerase II promoter | | nucleus | | negative regulation of DNA recombination | | transcription from RNA polymerase I promoter | | RNA polymerase II core binding | | positive regulation of histone H3-K36 trimethylation |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | Not Available |
|---|
| Locus | |
|---|
| Gene Sequence | >1338 bp
ATGTCCAAAAAACAGGAATATATTGCACCAATAAAATACCAAAACAGCTTGCCTGTGCCA
CAATTACCTCCTAAACTTCTGGTTTACCCCGAGTCTCCGGAGACGAATGCAGACTCCTCC
CAACTAATCAATTCACTATACATCAAAACAAACGTCACTAATCTAATTCAACAGGATGAA
GATTTAGGTATGCCGGTTGATTTGATGAAATTCCCAGGCCTATTGAACAAATTAGATTCT
AAACTGCTTTACGGCTTTGATAATGTGAAATTGGACAAAGATGATCGAATTTTACTGAGG
GACCCTAGAATAGATAGACTGACCAAGACTGATATATCAAAGGTTACCTTCTTGAGACGC
ACCGAATATGTCTCCAATACAATTGCAGCCCATGATAACACATCGTTGAAAAGGAAAAGG
CGCTTGGATGATGGAGATTCGGATGATGAAAACCTTGATGTTAATCATATAATTAGTAGG
GTCGAAGGAACATTCAATAAGACGGACAAATGGCAACATCCGGTGAAGAAGGGTGTCAAG
ATGGTTAAAAAATGGGACCTATTGCCCGACACAGCTTCAATGGACCAGGTGTATTTTATC
CTAAAATTCATGGGCAGTGCATCATTGGATACTAAAGAAAAAAAATCTTTAAATACAGGA
ATTTTCAGGCCAGTGGAACTGGAAGAAGACGAATGGATTTCCATGTATGCCACAGATCAT
AAGGACTCTGCCATTCTGGAAAACGAATTGGAAAAGGGTATGGATGAAATGGATGATGAC
TCTCATGAAGGTAAAATATACAAGTTTAAAAGGATAAGAGACTACGACATGAAACAAGTG
GCTGAAAAGCCAATGACAGAGTTAGCTATACGTTTGAATGATAAGGACGGCATAGCTTAC
TACAAACCCCTACGTTCAAAGATCGAATTAAGACGTAGGAGAGTAAACGACATCATTAAA
CCTTTGGTGAAGGAGCACGATATAGACCAACTTAATGTTACTTTGAGAAACCCTAGCACT
AAGGAAGCTAACATAAGGGACAAGTTAAGAATGAAGTTTGACCCCATAAATTTCGCCACT
GTAGATGAAGAAGACGATGAAGACGAGGAGCAACCAGAAGACGTTAAGAAAGAATCGGAA
GGCGATTCAAAAACAGAAGGTTCTGAGCAAGAAGGAGAAAATGAGAAGGATGAAGAAATC
AAACAGGAAAAAGAGAATGAACAAGATGAAGAAAATAAACAAGATGAAAACCGTGCAGCT
GATACGCCCGAAACTTCAGATGCTGTTCATACTGAACAAAAACCAGAGGAAGAAAAGGAA
ACTTTACAAGAAGAATAG |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | Not Available |
|---|
| Protein Residues | 445 |
|---|
| Protein Molecular Weight | 51800.4 |
|---|
| Protein Theoretical pI | Not Available |
|---|
| Signalling Regions | Not Available |
|---|
| Transmembrane Regions | Not Available |
|---|
| Protein Sequence | >RNA polymerase II-associated protein 1
MSKKQEYIAPIKYQNSLPVPQLPPKLLVYPESPETNADSSQLINSLYIKTNVTNLIQQDE
DLGMPVDLMKFPGLLNKLDSKLLYGFDNVKLDKDDRILLRDPRIDRLTKTDISKVTFLRR
TEYVSNTIAAHDNTSLKRKRRLDDGDSDDENLDVNHIISRVEGTFNKTDKWQHPVKKGVK
MVKKWDLLPDTASMDQVYFILKFMGSASLDTKEKKSLNTGIFRPVELEEDEWISMYATDH
KDSAILENELEKGMDEMDDDSHEGKIYKFKRIRDYDMKQVAEKPMTELAIRLNDKDGIAY
YKPLRSKIELRRRRVNDIIKPLVKEHDIDQLNVTLRNPSTKEANIRDKLRMKFDPINFAT
VDEEDDEDEEQPEDVKKESEGDSKTEGSEQEGENEKDEEIKQEKENEQDEENKQDENRAA
DTPETSDAVHTEQKPEEEKETLQEE |
|---|
| References |
|---|
| External Links | | Resource | Link |
|---|
| Saccharomyces Genome Database | PAF1 | | Uniprot ID | P38351 | | Uniprot Name | PAF1 |
|
|---|
| General Reference | Not Available |
|---|