| Identification |
|---|
| Name | Cell wall integrity sensor MID2 |
|---|
| Synonyms | - Mating pheromone-induced death protein 2
- Protein kinase A interference protein 1
- Serine-rich multicopy suppressor protein 1
|
|---|
| Gene Name | MID2 |
|---|
| Enzyme Class | Not Available |
|---|
| Biological Properties |
|---|
| General Function | response to osmotic stress |
|---|
| Specific Function | Cell wall stress sensor. Involved in activation of a response that includes both stress-related increased chitin synthesis and the MPK1 mitogen-activated protein kinase cell integrity pathway. High-copy activator of the SKN7 transcription factor. Required during induction of MPK1 tyrosine phosphorylation under a variety of stress conditions. Required for PKC pathway activation under low pH conditions. Required for cell integrity signaling during pheromone-induced morphogenesis. Activates ROM1 or ROM2 catalyzed guanine nucleotide exchange toward RHO1 in cell wall integrity signaling. |
|---|
| Cellular Location | Not Available |
|---|
| SMPDB Pathways | | Stress-activated signalling pathways: cell wall stress test 1 | PW007861 | 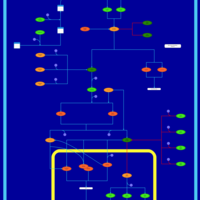 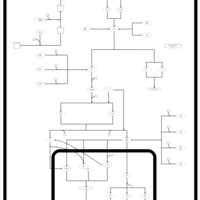 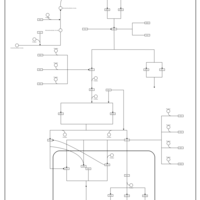 |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Metabolites | |
|---|
| GO Classification | | Function |
|---|
| integral component of plasma membrane | | fungal-type cell wall organization | | pexophagy | | response to osmotic stress | | transmembrane signaling receptor activity | | cell morphogenesis involved in conjugation | | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response | | response to acidic pH |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | Not Available |
|---|
| Locus | |
|---|
| Gene Sequence | >1131 bp
ATGTTGTCTTTCACAACCAAGAATAGTTTCCGCTTATTACTTTTAATACTGTCATGCATA
TCGACGATACGCGCACAATTTTTCGTGCAATCATCATCTTCGAATTCTTCAGCAGTATCT
ACTGCACGTTCTTCCGTAAGTAGAGTTAGTTCTTCAAGTTCCATTTTGTCATCCAGTATG
GTTTCTTCCTCAAGTGCTGACTCATCTTCCCTTACTTCATCGACATCAAGTAGGTCCCTC
GTGTCACATACGAGTTCGTCTACCAGCATTGCCTCCATATCGTTCACATCATTCAGTTTC
TCATCAGATTCAAGCACCAGCAGCTCTTCCTCTGCTTCTTCAGATTCTTCATCCTCCTCA
TCCTTTTCCATCTCTTCGACATCCGCAACTTCTGAATCATCGACGTCTTCTACGCAAACG
TCAACATCATCTTCATCCTCACTGTCGTCAACGCCGTCCTCTTCATCATCCCCATCAACA
ATCACTTCTGCACCTTCAACCTCCTCCACACCATCCACTACTGCCTATAATCAAGGAAGC
ACTATCACCAGTATTATTAACGGTAAGACGATTCTCTCAAATCATTATACTACCGTTACG
TACACACCATCAGCAACCGCTGATTCAAGCAATAAATCCAAAAGTTCGGGTCTTTCTAAA
AAAAATAGAAACATCGTCATTGGTTGTGTGGTTGGTATTGGTGTACCACTGATTCTGGTG
ATACTGGCTTTAATTTACATGTTTTGTATCCAATCATCAAGGACGGACTTTATTGACTCA
GATGGTAAGGTCGTTACGGCATATAGAGCTAACAAATTTACGAAATGGTGGTATATGCTT
TTGGGTAAGAAAGTTAGTGATGAATACCACTCCGATTCACCATTAGGTGGTAGTGCATCG
TCCGCAGGTGGTTTAGACCTAGATGAAGCAGATGATGTGATGGAGCAAAGCTCCCTTTTC
GACGTCAGAATAAGAGATAGCGATAGCGTATTACCAAACGCCAACACCGCCGACCATAAT
AATACCAATAGCGGTGGAGAGCCTATAAATAGTAGCGTTGCGTCAAATGACATAATAGAA
GAAAAATTCTATGATGAACAAGGTAACGAATTATCACCACGAAATTATTAA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | Not Available |
|---|
| Protein Residues | 376 |
|---|
| Protein Molecular Weight | 39145.835 |
|---|
| Protein Theoretical pI | Not Available |
|---|
| Signalling Regions | |
|---|
| Transmembrane Regions | |
|---|
| Protein Sequence | >Cell wall integrity sensor MID2
MLSFTTKNSFRLLLLILSCISTIRAQFFVQSSSSNSSAVSTARSSVSRVSSSSSILSSSM
VSSSSADSSSLTSSTSSRSLVSHTSSSTSIASISFTSFSFSSDSSTSSSSSASSDSSSSS
SFSISSTSATSESSTSSTQTSTSSSSSLSSTPSSSSSPSTITSAPSTSSTPSTTAYNQGS
TITSIINGKTILSNHYTTVTYTPSATADSSNKSKSSGLSKKNRNIVIGCVVGIGVPLILV
ILALIYMFCIQSSRTDFIDSDGKVVTAYRANKFTKWWYMLLGKKVSDEYHSDSPLGGSAS
SAGGLDLDEADDVMEQSSLFDVRIRDSDSVLPNANTADHNNTNSGGEPINSSVASNDIIE
EKFYDEQGNELSPRNY |
|---|
| References |
|---|
| External Links | | Resource | Link |
|---|
| Saccharomyces Genome Database | MID2 | | Uniprot ID | P36027 | | Uniprot Name | MID2 |
|
|---|
| General Reference | Not Available |
|---|