| Identification |
|---|
| Name | G2/mitotic-specific cyclin-1 |
|---|
| Synonyms | Not Available |
|---|
| Gene Name | CLB1 |
|---|
| Enzyme Class | Not Available |
|---|
| Biological Properties |
|---|
| General Function | regulation of cyclin-dependent protein serine/threonine kinase activity |
|---|
| Specific Function | Essential for the control of the cell cycle at the G2/M (mitosis) transition. Interacts with the CDC2 protein kinase to form MPF. G2/M cyclins accumulate steadily during G2 and are abruptly destroyed at mitosis. |
|---|
| Cellular Location | Not Available |
|---|
| SMPDB Pathways | | Stress-activated signalling pathways: cell wall stress test 1 | PW007861 | 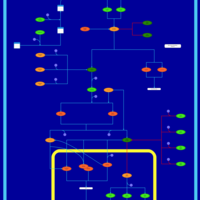 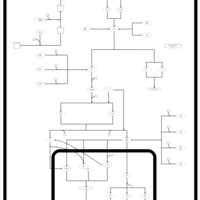 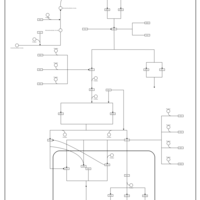 | | Stress-activated signalling pathways: high osmolarity test 1 | PW002773 | 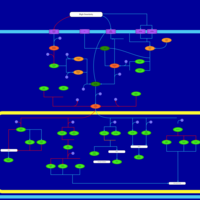 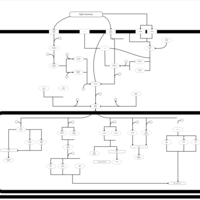 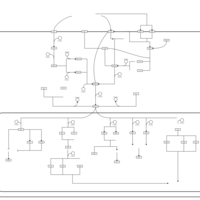 |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Metabolites | |
|---|
| GO Classification | | Function |
|---|
| meiotic G2/MI transition | | mitotic spindle organization in nucleus | | nucleus | | cyclin-dependent protein kinase holoenzyme complex | | cytoplasm | | cell division | | positive regulation of spindle pole body separation | | mitotic nuclear division | | cyclin-dependent protein serine/threonine kinase regulator activity | | regulation of cyclin-dependent protein serine/threonine kinase activity | | G2/M transition of mitotic cell cycle |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | Not Available |
|---|
| Locus | |
|---|
| Gene Sequence | >1416 bp
ATGTCACGATCCCTTTTGGTAGAGAATAGTAGAACCATTAATAGTAATGAAGAGAAGGGG
GTAAACGAAAGTCAATATATCTTACAAAAGAGAAACGTCCCAAGGACCATTCTCGGTAAT
GTCACAAATAATGCAAATATCCTGCAAGAGATTTCTATGAACAGAAAAATTGGGATGAAG
AACTTCTCAAAATTGAACAACTTCTTTCCTTTAAAGGATGATGTTTCGAGAGCCGATGAC
TTCACCTCCTCTTTTAATGACAGTAGACAGGGGGTGAAACAAGAAGTACTAAACAATAAG
GAAAATATTCCTGAATATGGATATTCCGAGCAAGAAAAGCAGCAGTGCTCAAATGATGAC
TCTTTTCATACGAATTCGACTGCCTTGAGCTGTAATCGGTTGATATACTCTGAGAACAAG
TCTATTTCCACTCAAATGGAATGGCAGAAAAAAATAATGAGAGAAGATTCCAAAAAAAAA
AGGCCAATATCAACTCTTGTAGAGCAGGATGACCAGAAAAAGTTCAAACTCCATGAATTG
ACTACAGAAGAAGAGGTTCTGGAGGAGTACGAATGGGATGACCTAGATGAAGAAGATTGT
GATGATCCCTTAATGGTGAGCGAAGAGGTTAATGATATCTTCGATTATTTGCATCACTTG
GAAATAATCACATTGCCAAATAAGGCGAATCTCTATAAGCATAAAAATATTAAGCAAAAC
AGAGACATTTTGGTGAACTGGATAATCAAGATTCATAATAAGTTTGGCCTTCTGCCGGAA
ACTCTATATTTGGCCATAAACATAATGGATAGATTTCTTTGTGAAGAGGTGGTTCAGTTA
AATAGATTGCAACTGGTTGGTACATCATGCCTGTTCATCGCATCTAAGTATGAGGAAATA
TACTCCCCTAGCATAAAACATTTTGCGTACGAGACAGACGGTGCATGCTCTGTGGAAGAT
ATTAAAGAGGGTGAAAGATTTATATTGGAAAAACTCGATTTTCAAATTAGTTTCGCTAAC
CCAATGAATTTCTTAAGGAGAATATCAAAAGCGGATGACTATGATATCCAGTCTAGGACG
TTAGCGAAGTTCCTAATGGAAATATCTATAGTGGACTTCAAGTTCATCGGGATATTACCG
TCATTATGTGCCTCAGCGGCAATGTTCCTTTCGAGAAAAATGCTGGGCAAGGGCACCTGG
GATGGCAACTTAATCCATTATAGTGGTGGTTACACAAAGGCAAAACTATATCCCGTTTGC
CAATTACTGATGGATTATCTTGTTGGATCTACTATTCACGACGAGTTCTTGAAAAAATAT
CAATCGAGAAGATTTTTAAAGGCTTCCATAATTTCTATCGAATGGGCATTGAAGGTAAGG
AAAAATGGATATGATATTATGACATTGCATGAGTGA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | Not Available |
|---|
| Protein Residues | 471 |
|---|
| Protein Molecular Weight | 54869.99 |
|---|
| Protein Theoretical pI | Not Available |
|---|
| Signalling Regions | Not Available |
|---|
| Transmembrane Regions | Not Available |
|---|
| Protein Sequence | >G2/mitotic-specific cyclin-1
MSRSLLVENSRTINSNEEKGVNESQYILQKRNVPRTILGNVTNNANILQEISMNRKIGMK
NFSKLNNFFPLKDDVSRADDFTSSFNDSRQGVKQEVLNNKENIPEYGYSEQEKQQCSNDD
SFHTNSTALSCNRLIYSENKSISTQMEWQKKIMREDSKKKRPISTLVEQDDQKKFKLHEL
TTEEEVLEEYEWDDLDEEDCDDPLMVSEEVNDIFDYLHHLEIITLPNKANLYKHKNIKQN
RDILVNWIIKIHNKFGLLPETLYLAINIMDRFLCEEVVQLNRLQLVGTSCLFIASKYEEI
YSPSIKHFAYETDGACSVEDIKEGERFILEKLDFQISFANPMNFLRRISKADDYDIQSRT
LAKFLMEISIVDFKFIGILPSLCASAAMFLSRKMLGKGTWDGNLIHYSGGYTKAKLYPVC
QLLMDYLVGSTIHDEFLKKYQSRRFLKASIISIEWALKVRKNGYDIMTLHE |
|---|
| References |
|---|
| External Links | | Resource | Link |
|---|
| Saccharomyces Genome Database | CLB1 | | Uniprot ID | P24868 | | Uniprot Name | CLB1 |
|
|---|
| General Reference | Not Available |
|---|