| Identification |
|---|
| Name | Mitogen-activated protein kinase FUS3 |
|---|
| Synonyms | |
|---|
| Gene Name | FUS3 |
|---|
| Enzyme Class | Not Available |
|---|
| Biological Properties |
|---|
| General Function | protein phosphorylation |
|---|
| Specific Function | Together with closely related KSS1, FUS3 is the final kinase in the signal transduction cascade regulating activation/repression of the mating and filamentation pathways, induced by pheromone and nitrogen/carbon limitation, respectively. Phosphorylated FUS3 activates the mating but suppresses the filamentation pathway, whereas activated KSS1 activates both pathways. Pheromone-activated FUS3 functions by inhibiting the binding of the transcriptional activator STE12 to filamentation specific genes while inducing its binding to and activity at mating specific genes. Non-activated FUS3 has a repressive effect on STE12 transcriptional activity. KSS1 can partially compensate for the lack of FUS3 but mating efficiency is reduced and the filamentation program is partially activated upon pheromone signaling. FUS3 phosphorylates STE7, STE5, FAR1, DIG1, DIG2 and STE12. |
|---|
| Cellular Location | Not Available |
|---|
| SMPDB Pathways | | Stress-activated signalling pathways: pheromone stress test 1 | PW012843 |  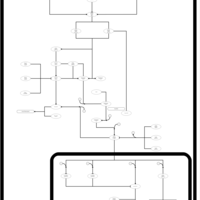 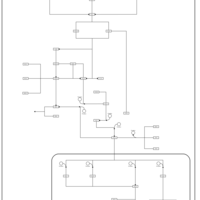 |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Metabolites | |
|---|
| GO Classification | | Function |
|---|
| cytoplasm | | cell division | | mating projection tip | | invasive growth in response to glucose limitation | | protein phosphorylation | | periplasmic space | | MAP kinase activity | | identical protein binding | | cell cycle arrest | | mitotic nuclear division | | negative regulation of MAPK cascade | | negative regulation of transposition, RNA-mediated | | positive regulation of protein export from nucleus | | ATP binding | | protein autophosphorylation | | pheromone-dependent signal transduction involved in conjugation with cellular fusion | | nucleus |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | Not Available |
|---|
| Locus | |
|---|
| Gene Sequence | >1062 bp
ATGCCAAAGAGAATTGTATACAATATATCCAGTGACTTCCAGTTGAAGTCGTTACTGGGA
GAGGGTGCATACGGTGTGGTATGTTCTGCAACGCATAAGCCCACGGGAGAAATCGTGGCA
ATAAAAAAGATCGAACCATTCGATAAGCCTTTGTTCGCATTACGTACGCTGCGTGAAATA
AAGATCCTGAAGCACTTCAAGCACGAAAATATCATAACAATCTTCAACATTCAACGCCCT
GACTCGTTCGAAAACTTCAATGAGGTCTACATAATTCAAGAGCTAATGCAGACAGATTTA
CACCGTGTAATCTCCACCCAGATGCTGAGTGACGATCATATACAATATTTTATATACCAA
ACCTTGAGAGCAGTGAAAGTGCTGCATGGTTCGAACGTCATCCATCGTGATTTAAAGCCC
TCCAACCTTCTCATAAACTCCAACTGTGACTTGAAAGTATGTGATTTCGGTTTAGCAAGA
ATCATTGACGAGTCAGCCGCGGACAATTCAGAGCCCACAGGTCAGCAAAGCGGCATGACC
GAGTATGTGGCCACACGTTGGTACAGGGCGCCAGAGGTGATGTTAACCTCTGCCAAATAC
TCAAGGGCCATGGACGTGTGGTCCTGCGGATGTATTCTCGCTGAACTTTTCTTAAGACGG
CCAATCTTCCCTGGCAGAGATTATCGCCATCAACTACTACTGATATTCGGTATCATCGGT
ACACCTCACTCAGATAATGATTTGCGGTGTATAGAGTCACCCAGGGCTAGAGAGTACATA
AAGTCGCTTCCCATGTACCCTGCCGCGCCACTGGAGAAGATGTTCCCTCGAGTCAACCCG
AAAGGCATAGATCTTTTACAGCGTATGCTTGTTTTTGACCCTGCGAAGAGGATTACTGCT
AAGGAGGCACTGGAGCATCCGTATTTGCAAACATACCACGATCCAAACGACGAACCTGAA
GGCGAACCCATCCCACCCAGCTTCTTCGAGTTTGATCACTACAAGGAGGCACTAACGACG
AAAGACCTCAAGAAACTCATTTGGAACGAAATATTTAGTTAG |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | Not Available |
|---|
| Protein Residues | 353 |
|---|
| Protein Molecular Weight | 40771.63 |
|---|
| Protein Theoretical pI | Not Available |
|---|
| PDB File | show |
|---|
| Signalling Regions | Not Available |
|---|
| Transmembrane Regions | Not Available |
|---|
| Protein Sequence | >Mitogen-activated protein kinase FUS3
MPKRIVYNISSDFQLKSLLGEGAYGVVCSATHKPTGEIVAIKKIEPFDKPLFALRTLREI
KILKHFKHENIITIFNIQRPDSFENFNEVYIIQELMQTDLHRVISTQMLSDDHIQYFIYQ
TLRAVKVLHGSNVIHRDLKPSNLLINSNCDLKVCDFGLARIIDESAADNSEPTGQQSGMT
EYVATRWYRAPEVMLTSAKYSRAMDVWSCGCILAELFLRRPIFPGRDYRHQLLLIFGIIG
TPHSDNDLRCIESPRAREYIKSLPMYPAAPLEKMFPRVNPKGIDLLQRMLVFDPAKRITA
KEALEHPYLQTYHDPNDEPEGEPIPPSFFEFDHYKEALTTKDLKKLIWNEIFS |
|---|
| References |
|---|
| External Links | | Resource | Link |
|---|
| Saccharomyces Genome Database | FUS3 | | Uniprot ID | P16892 | | Uniprot Name | FUS3 | | PDB ID | 2B9F |
|
|---|
| General Reference | Not Available |
|---|