| Identification |
|---|
| Name | Protein UPS1, mitochondrial |
|---|
| Synonyms | - Unprocessed MGM1 protein 1
|
|---|
| Gene Name | UPS1 |
|---|
| Enzyme Class | Not Available |
|---|
| Biological Properties |
|---|
| General Function | positive regulation of phosphatidylcholine biosynthetic process |
|---|
| Specific Function | Required for maintenance of normal mitochondrial morphology (PubMed:16754953, PubMed:26071601). Required for PCP1-dependent processing of MGM1 (PubMed:16754953). The UPS1:MDM35 complex mediates the transfer of phosphatidic acid (PA) between liposomes and probably functions as a PA transporter across the mitochondrion intermembrane space (PubMed:26071602, PubMed:26071601, PubMed:26235513). Phosphatidic acid release requires dissociation of the UPS1:MDM35 complex (PubMed:26235513). Phosphatidic acid import is required for cardiolipin (CL) synthesis in the mitochondrial inner membrane (PubMed:26071602). With UPS2, controls the level of cardiolipin in mitochondria (PubMed:19506038). Cardiolipin is a unique phospholipid with four fatty acid chains and is present mainly in the mitochondrial inner membrane where it stabilizes the electron transport chain supercomplex between complexes III and IV through direct interaction of their subunits. |
|---|
| Cellular Location | Not Available |
|---|
| SMPDB Pathways | | Cardiolipin Biosynthesis | PW002431 | 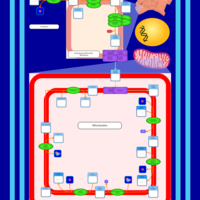 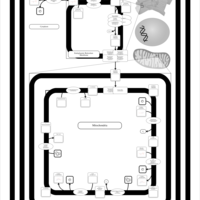 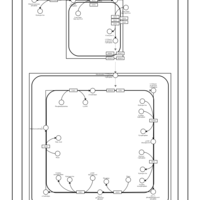 | | Cardiolipin Biosynthesis CL(10:0/10:0/10:0/28:0) | PW003484 | 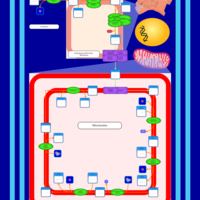 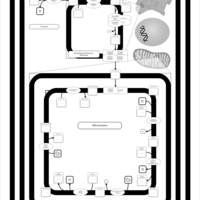 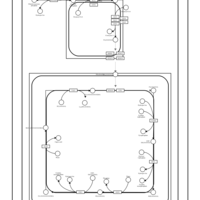 | | Cardiolipin Biosynthesis CL(10:0/10:0/10:0/30:0) | PW003501 | 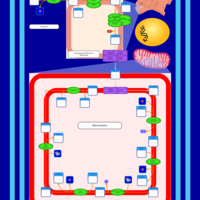 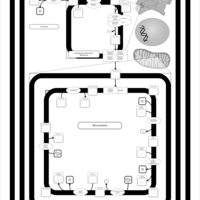 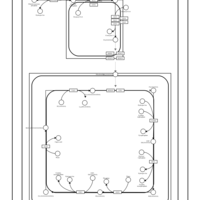 | | Cardiolipin Biosynthesis CL(10:0/10:0/12:0/26:0) | PW003502 | 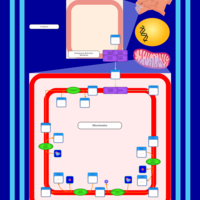 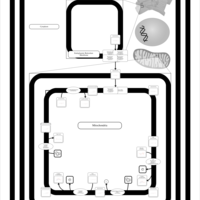 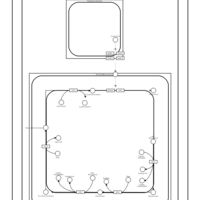 | | Cardiolipin Biosynthesis CL(10:0/10:0/12:0/28:0) | PW003503 | 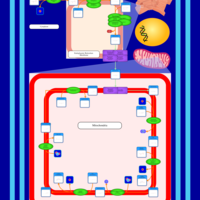 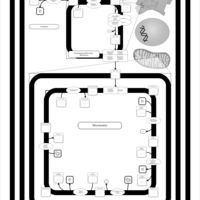 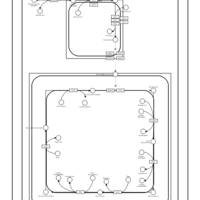 |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Metabolites | |
|---|
| GO Classification | | Function |
|---|
| phospholipid transport | | extrinsic component of mitochondrial inner membrane | | phosphatidic acid transporter activity | | cardiolipin metabolic process | | positive regulation of phosphatidylcholine biosynthetic process | | lipid binding | | mitochondrial intermembrane space |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | Not Available |
|---|
| Locus | |
|---|
| Gene Sequence | >528 bp
ATGGTCCTTTTACACAAAAGCACACATATATTTCCTACCGATTTTGCCTCTGTTTCACGC
GCCTTTTTTAATAGATACCCCAATCCATACTCCCCCCATGTACTATCCATAGACACAATA
TCAAGGAACGTTGATCAAGAAGGAAATTTGCGCACAACGAGGCTGTTGAAAAAGTCCGGA
AAGCTGCCCACATGGGTCAAACCCTTTTTAAGAGGTATAACAGAAACATGGATAATCGAA
GTTTCCGTAGTGAACCCCGCTAACTCCACAATGAAAACTTACACTAGGAATCTGGATCAC
ACTGGAATCATGAAGGTTGAAGAATATACTACCTATCAATTTGACAGTGCTACAAGTAGT
ACGATAGCAGACAGCCGGGTGAAGTTTTCAAGTGGCTTCAATATGGGTATCAAATCTAAG
GTAGAGGATTGGTCGCGCACTAAATTTGACGAAAACGTTAAGAAAAGCAGAATGGGCATG
GCATTTGTTATCCAAAAACTCGAAGAGGCGAGAAATCCTCAGTTTTGA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | Not Available |
|---|
| Protein Residues | 175 |
|---|
| Protein Molecular Weight | 20107.82 |
|---|
| Protein Theoretical pI | Not Available |
|---|
| PDB File | show |
|---|
| Signalling Regions | Not Available |
|---|
| Transmembrane Regions | Not Available |
|---|
| Protein Sequence | >Protein UPS1, mitochondrial
MVLLHKSTHIFPTDFASVSRAFFNRYPNPYSPHVLSIDTISRNVDQEGNLRTTRLLKKSG
KLPTWVKPFLRGITETWIIEVSVVNPANSTMKTYTRNLDHTGIMKVEEYTTYQFDSATSS
TIADSRVKFSSGFNMGIKSKVEDWSRTKFDENVKKSRMGMAFVIQKLEEARNPQF |
|---|
| References |
|---|
| External Links | | Resource | Link |
|---|
| Saccharomyces Genome Database | UPS1 | | Uniprot ID | Q05776 | | Uniprot Name | UPS1 | | PDB ID | 4XHR |
|
|---|
| General Reference | Not Available |
|---|