| Identification |
|---|
| Name | Biotin--protein ligase |
|---|
| Synonyms | - Biotin apo-protein ligase
- Biotin--[methylmalonyl-CoA-carboxytransferase] ligase
- Biotin--[propionyl-CoA-carboxylase [ATP-hydrolyzing]] ligase
- Holocarboxylase synthetase
- HCS
- Biotin--[methylcrotonoyl-CoA-carboxylase] ligase
- Biotin--[acetyl-CoA-carboxylase] ligase
|
|---|
| Gene Name | BPL1 |
|---|
| Enzyme Class | |
|---|
| Biological Properties |
|---|
| General Function | Involved in biotin-[acetyl-CoA-carboxylase] ligase activity |
|---|
| Specific Function | Post-translational modification of specific protein by attachment of biotin. Acts on various carboxylases such as acetyl- CoA-carboxylase, pyruvate carboxylase, propionyl CoA carboxylase, and 3-methylcrotonyl CoA carboxylase |
|---|
| Cellular Location | Cytoplasm |
|---|
| SMPDB Pathways | Not Available |
|---|
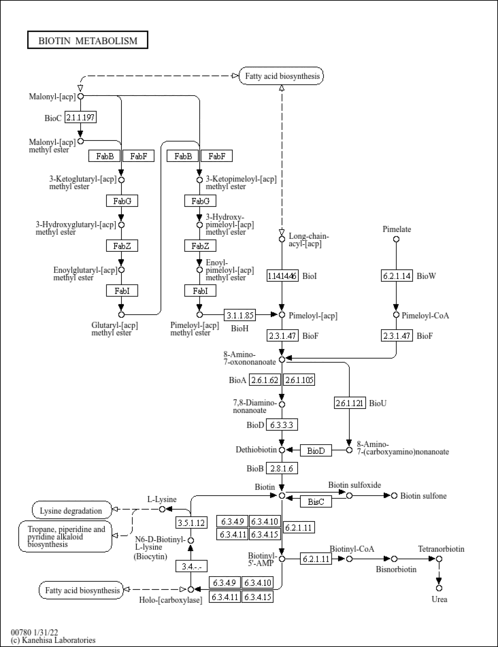
| KEGG Pathways | |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | |
|---|
| Metabolites | | YMDB ID | Name | View |
|---|
| YMDB00097 | Adenosine monophosphate | Show | | YMDB00109 | Adenosine triphosphate | Show | | YMDB00219 | Pyrophosphate | Show | | YMDB00282 | Biotin | Show | | YMDB00602 | Biotinyl-5'-AMP | Show | | YMDB00862 | hydron | Show |
|
|---|
| GO Classification | | Component |
|---|
| Not Available | | Function |
|---|
| ligase activity | | ligase activity, forming carbon-nitrogen bonds | | biotin-protein ligase activity | | biotin-[acetyl-CoA-carboxylase] ligase activity | | catalytic activity | | Process |
|---|
| metabolic process | | macromolecule metabolic process | | macromolecule modification | | protein modification process |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | chromosome 4 |
|---|
| Locus | YDL141W |
|---|
| Gene Sequence | >2073 bp
ATGAATGTATTAGTCTATAATGGCCCAGGGACTACGCCAGGATCAGTTAAGCATGCTGTT
GAAAGTTTACGTGACTTTTTGGAACCCTATTACGCCGTCAGTACTGTCAATGTTAAGGTA
TTACAGACAGAACCATGGATGTCAAAGACCTCCGCTGTAGTTTTTCCTGGTGGTGCCGAT
CTTCCTTACGTTCAAGCTTGTCAACCCATTATTTCTCGATTGAAACATTTTGTTAGCAAA
CAAGGTGGTGTCTTTATCGGATTTTGTGCAGGAGGTTACTTTGGCACCAGCCGTGTAGAA
TTTGCTCAAGGTGACCCTACAATGGAAGTATCAGGAAGCAGGGATTTGCGATTCTTTCCG
GGAACAAGCAGAGGACCTGCGTACAATGGATTTCAGTATAACAGCGAGGCCGGTGCACGT
GCAGTTAAATTGAATCTTCCAGATGGCTCACAGTTCTCAACTTATTTCAATGGAGGAGCC
GTTTTTGTTGATGCAGATAAATTTGATAATGTCGAGATCTTGGCCACGTACGCAGAACAC
CCGGATGTGCCTTCTTCTGACTCAGGAAAAGGTCAAAGTGAAAACCCAGCTGCTGTAGTG
CTATGTACGGTCGGAAGAGGAAAGGTTTTACTCACTGGACCACACCCTGAATTCAACGTC
CGTTTTATGAGGAAATCCACAGATAAGCATTTTCTTGAAACCGTTGTGGAAAACCTGAAA
GCACAGGAAATCATGAGGTTGAAATTTATGAGAACGGTTCTTACCAAAACTGGGTTGAAT
TGTAATAATGACTTCAATTATGTAAGAGCTCCAAATTTGACACCACTGTTTATGGCTAGT
GCACCTAATAAGCGGAATTACTTGCAAGAAATGGAAAACAACTTAGCTCATCATGGAATG
CATGCAAATAATGTTGAACTCTGCTCAGAATTAAACGCCGAAACTGACTCTTTCCAATTT
TATAGAGGCTATAGAGCCTCTTACGATGCAGCAAGTTCGTCCTTGCTTCATAAAGAGCCA
GATGAGGTTCCTAAGACCGTAATATTTCCTGGCGTAGATGAAGATATTCCACCATTTCAA
TATACGCCCAATTTTGACATGAAAGAATACTTCAAGTATCTGAATGTTCAAAACACCATA
GGTTCTTTACTTCTATATGGTGAAGTTGTTACATCAACGAGTACCATATTGAATAATAAT
AAATCCTTATTAAGTTCGATCCCTGAAAGCACTTTACTTCACGTGGGTACTATTCAGGTT
TCTGGGCGTGGGAGAGGTGGTAACACTTGGATTAATCCTAAAGGTGTTTGCGCATCAACG
GCCGTCGTAACAATGCCGCTTCAATCGCCTGTGACTAATCGAAATATATCAGTTGTTTTT
GTCCAGTATCTTTCTATGCTTGCTTACTGTAAGGCGATACTTTCGTACGCTCCTGGATTT
TCAGATATTCCTGTTAGGATCAAATGGCCCAATGACCTTTATGCTTTGAGCCCCACCTAC
TATAAGCGTAAAAACTTGAAATTGGTTAACACCGGTTTTGAGCATACAAAGTTGCCCTTA
GGTGATATCGAGCCCGCTTATCTTAAGATCTCTGGTTTATTAGTTAATACCCACTTCATA
AACAATAAATATTGTTTGCTACTTGGCTGTGGTATAAATTTGACTAGTGATGGTCCTACA
ACTTCATTACAAACTTGGATTGATATCCTGAATGAAGAACGCCAACAATTACACCTTGAT
TTATTGCCAGCAATAAAGGCAGAAAAGCTGCAGGCACTCTATATGAATAACCTTGAGGTC
ATATTAAAACAGTTTATTAACTACGGAGCTGCGGAAATATTGCCAAGTTATTATGAGTTG
TGGTTACATAGTAACCAAATAGTAACATTACCAGATCACGGAAATACGCAAGCCATGATT
ACTGGTATCACGGAAGACTACGGTTTACTGATTGCAAAAGAATTGGTAAGCGGTAGTTCC
ACTCAATTTACAGGAAACGTTTATAATTTACAGCCGGACGGTAACACTTTTGATATCTTC
AAGAGCTTAATTGCTAAAAAGGTTCAGAGTTAA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | |
|---|
| Protein Residues | 690 |
|---|
| Protein Molecular Weight | 76362.39844 |
|---|
| Protein Theoretical pI | 6.91 |
|---|
| Signalling Regions | |
|---|
| Transmembrane Regions | |
|---|
| Protein Sequence | >Biotin--protein ligase
MNVLVYNGPGTTPGSVKHAVESLRDFLEPYYAVSTVNVKVLQTEPWMSKTSAVVFPGGAD
LPYVQACQPIISRLKHFVSKQGGVFIGFCAGGYFGTSRVEFAQGDPTMEVSGSRDLRFFP
GTSRGPAYNGFQYNSEAGARAVKLNLPDGSQFSTYFNGGAVFVDADKFDNVEILATYAEH
PDVPSSDSGKGQSENPAAVVLCTVGRGKVLLTGPHPEFNVRFMRKSTDKHFLETVVENLK
AQEIMRLKFMRTVLTKTGLNCNNDFNYVRAPNLTPLFMASAPNKRNYLQEMENNLAHHGM
HANNVELCSELNAETDSFQFYRGYRASYDAASSSLLHKEPDEVPKTVIFPGVDEDIPPFQ
YTPNFDMKEYFKYLNVQNTIGSLLLYGEVVTSTSTILNNNKSLLSSIPESTLLHVGTIQV
SGRGRGGNTWINPKGVCASTAVVTMPLQSPVTNRNISVVFVQYLSMLAYCKAILSYAPGF
SDIPVRIKWPNDLYALSPTYYKRKNLKLVNTGFEHTKLPLGDIEPAYLKISGLLVNTHFI
NNKYCLLLGCGINLTSDGPTTSLQTWIDILNEERQQLHLDLLPAIKAEKLQALYMNNLEV
ILKQFINYGAAEILPSYYELWLHSNQIVTLPDHGNTQAMITGITEDYGLLIAKELVSGSS
TQFTGNVYNLQPDGNTFDIFKSLIAKKVQS |
|---|
| References |
|---|
| External Links | |
|---|
| General Reference | - Cronan, J. E. Jr, Wallace, J. C. (1995). "The gene encoding the biotin-apoprotein ligase of Saccharomyces cerevisiae." FEMS Microbiol Lett 130:221-229.7649444
- Wolfl, S., Hanemann, V., Saluz, H. P. (1996). "Analysis of a 26,756 bp segment from the left arm of yeast chromosome IV." Yeast 12:1549-1554.8972577
- Jacq, C., Alt-Morbe, J., Andre, B., Arnold, W., Bahr, A., Ballesta, J. P., Bargues, M., Baron, L., Becker, A., Biteau, N., Blocker, H., Blugeon, C., Boskovic, J., Brandt, P., Bruckner, M., Buitrago, M. J., Coster, F., Delaveau, T., del Rey, F., Dujon, B., Eide, L. G., Garcia-Cantalejo, J. M., Goffeau, A., Gomez-Peris, A., Zaccaria, P., et, a. l. .. (1997). "The nucleotide sequence of Saccharomyces cerevisiae chromosome IV." Nature 387:75-78.9169867
- Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., O'Shea, E. K. (2003). "Global analysis of protein localization in budding yeast." Nature 425:686-691.14562095
- Ghaemmaghami, S., Huh, W. K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., O'Shea, E. K., Weissman, J. S. (2003). "Global analysis of protein expression in yeast." Nature 425:737-741.14562106
|
|---|