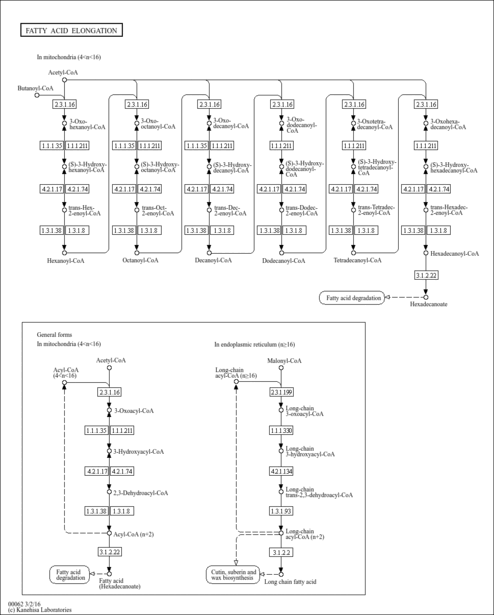
| General Reference | - Einerhand, A. W., Voorn-Brouwer, T. M., Erdmann, R., Kunau, W. H., Tabak, H. F. (1991). "Regulation of transcription of the gene coding for peroxisomal 3-oxoacyl-CoA thiolase of Saccharomyces cerevisiae." Eur J Biochem 200:113-122.1715273
- Igual, J. C., Matallana, E., Gonzalez-Bosch, C., Franco, L., Perez-Ortin, J. E. (1991). "A new glucose-repressible gene identified from the analysis of chromatin structure in deletion mutants of yeast SUC2 locus." Yeast 7:379-389.1872029
- Churcher, C., Bowman, S., Badcock, K., Bankier, A., Brown, D., Chillingworth, T., Connor, R., Devlin, K., Gentles, S., Hamlin, N., Harris, D., Horsnell, T., Hunt, S., Jagels, K., Jones, M., Lye, G., Moule, S., Odell, C., Pearson, D., Rajandream, M., Rice, P., Rowley, N., Skelton, J., Smith, V., Barrell, B., et, a. l. .. (1997). "The nucleotide sequence of Saccharomyces cerevisiae chromosome IX." Nature 387:84-87.9169870
- Hu, Y., Rolfs, A., Bhullar, B., Murthy, T. V., Zhu, C., Berger, M. F., Camargo, A. A., Kelley, F., McCarron, S., Jepson, D., Richardson, A., Raphael, J., Moreira, D., Taycher, E., Zuo, D., Mohr, S., Kane, M. F., Williamson, J., Simpson, A., Bulyk, M. L., Harlow, E., Marsischky, G., Kolodner, R. D., LaBaer, J. (2007). "Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae." Genome Res 17:536-543.17322287
- Glover, J. R., Andrews, D. W., Subramani, S., Rachubinski, R. A. (1994). "Mutagenesis of the amino targeting signal of Saccharomyces cerevisiae 3-ketoacyl-CoA thiolase reveals conserved amino acids required for import into peroxisomes in vivo." J Biol Chem 269:7558-7563.8125978
- Mathieu, M., Zeelen, J. P., Pauptit, R. A., Erdmann, R., Kunau, W. H., Wierenga, R. K. (1994). "The 2.8 A crystal structure of peroxisomal 3-ketoacyl-CoA thiolase of Saccharomyces cerevisiae: a five-layered alpha beta alpha beta alpha structure constructed from two core domains of identical topology." Structure 2:797-808.7812714
- Mathieu, M., Modis, Y., Zeelen, J. P., Engel, C. K., Abagyan, R. A., Ahlberg, A., Rasmussen, B., Lamzin, V. S., Kunau, W. H., Wierenga, R. K. (1997). "The 1.8 A crystal structure of the dimeric peroxisomal 3-ketoacyl-CoA thiolase of Saccharomyces cerevisiae: implications for substrate binding and reaction mechanism." J Mol Biol 273:714-728.9402066
|
|---|