Lysine biosynthesis
| Source | ID | Pathway |
|---|---|---|
| KEGG | 00300 | 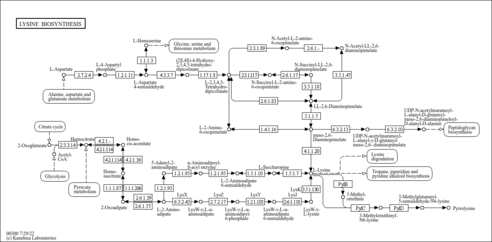 |
Pathway Metabolites
| YMDB ID | Name CAS Number | IUPAC Name | Formula Weight | Structure |
|---|---|---|---|---|
| YMDB00019 | Saccharopine 997-68-2 | (2S)-2-{[(5S)-5-amino-5-carboxypentyl]amino}pentanedioic acid | C11H20N2O6 276.2863 | 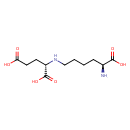 |
| YMDB00034 | L-aspartic 4-semialdehyde 15106-57-7 | (2S)-2-amino-4-oxobutanoic acid | C4H7NO3 117.1033 | 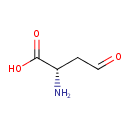 |
| YMDB00096 | (2R)-Homocitric acid 3562-74-1 | (2R)-2-hydroxybutane-1,2,4-tricarboxylic acid | C7H10O7 206.1501 | 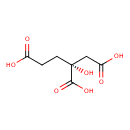 |
| YMDB00104 | L-Homoserine 672-15-1 | (2S)-2-amino-4-hydroxybutanoic acid | C4H9NO3 119.1192 | 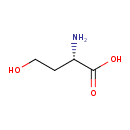 |
| YMDB00113 | 4-Phospho-L-aspartic acid 22138-53-0 | (2S)-2-amino-4-oxo-4-(phosphonooxy)butanoic acid | C4H8NO7P 213.0826 | 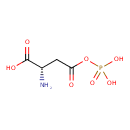 |
| YMDB00142 | Oxoadipic acid 3184-35-8 | 2-oxohexanedioic acid | C6H8O5 160.1247 | 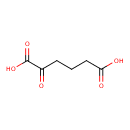 |
| YMDB00144 | Homoisocitric acid | (1R,2S)-1-hydroxybutane-1,2,4-tricarboxylic acid | C7H10O7 206.1501 | 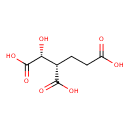 |
| YMDB00153 | Oxoglutaric acid 328-50-7 | 2-oxopentanedioic acid | C5H6O5 146.0981 | 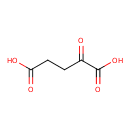 |
| YMDB00269 | but-1-ene-1,2,4-tricarboxylic acid | (1Z)-but-1-ene-1,2,4-tricarboxylic acid | C7H8O6 188.1348 | 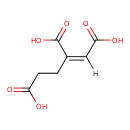 |
| YMDB00305 | Aminoadipic acid 542-32-5 | C6H11NO4 161.1558 | 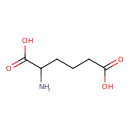 | |
| YMDB00312 | Acetyl-CoA 72-89-9 | {[(2R,3S,4R,5R)-2-({[({[(3R)-3-[(2-{[2-(acetylsulfanyl)ethyl]carbamoyl}ethyl)carbamoyl]-3-hydroxy-2,2-dimethylpropoxy](hydroxy)phosphoryl}oxy)(hydroxy)phosphoryl]oxy}methyl)-5-(6-amino-9H-purin-9-yl)-4-hydroxyoxolan-3-yl]oxy}phosphonic acid | C23H38N7O17P3S 809.571 | 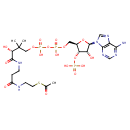 |
| YMDB00330 | L-Lysine 56-87-1 | (2S)-2,6-diaminohexanoic acid | C6H14N2O2 146.1876 | 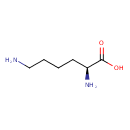 |
| YMDB00705 | 2-Oxaloglutaric acid | 1-oxobutane-1,2,4-tricarboxylic acid | C7H8O7 204.1342 | 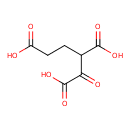 |
| YMDB00721 | L-Allysine 1962-83-0 | (2S)-2-amino-6-oxohexanoic acid | C6H11NO3 145.1564 | 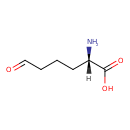 |
| YMDB00896 | L-Aspartic acid 56-84-8 | (2S)-2-aminobutanedioic acid | C4H7NO4 133.1027 | 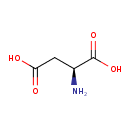 |
| YMDB00999 | L-2-aminoadipate 542-32-5 | (2S)-2-aminohexanedioic acid | C6H11NO4 161.1558 | 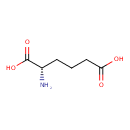 |
| YMDB01022 | L-aspartate 4-semialdehyde 15106-57-7 | C4H6NO3 116.0953 | 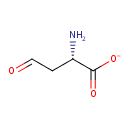 | |
| YMDB01067 | 4-phospho-L-aspartate 22138-53-0 | C4H5NO7P 210.0588 | 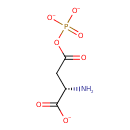 |
Pathway Proteins
| Uniprot ID | Gene Name Locus | Name | Type | Metabolites |
|---|---|---|---|---|
| P38998 | LYS1 YIR034C | Saccharopine dehydrogenase [NAD+, L-lysine-forming] | Enzyme | |
| P38999 | LYS9 YNR050C | Saccharopine dehydrogenase [NADP+, L-glutamate-forming] | Enzyme | |
| P18544 | ARG8 YOL140W | Acetylornithine aminotransferase, mitochondrial | Enzyme | |
| P13663 | HOM2 YDR158W | Aspartate-semialdehyde dehydrogenase | Enzyme | |
| Q12122 | LYS21 YDL131W | Homocitrate synthase, mitochondrial | Enzyme | |
| P48570 | LYS20 YDL182W | Homocitrate synthase, cytosolic isozyme | Enzyme | |
| P07702 | LYS2 YBR115C | L-aminoadipate-semialdehyde dehydrogenase | Enzyme | |
| P10869 | HOM3 YER052C | Aspartokinase | Enzyme | |
| P40495 | LYS12 YIL094C | Homoisocitrate dehydrogenase, mitochondrial | Enzyme | |
| P38840 | ARO9 YHR137W | Aromatic amino acid aminotransferase 2 | Enzyme | |
| P53090 | ARO8 YGL202W | Aromatic amino acid aminotransferase 1 | Enzyme | |
| P49367 | LYS4 YDR234W | Homoaconitase, mitochondrial | Enzyme | |
| P31116 | HOM6 YJR139C | Homoserine dehydrogenase | Enzyme |